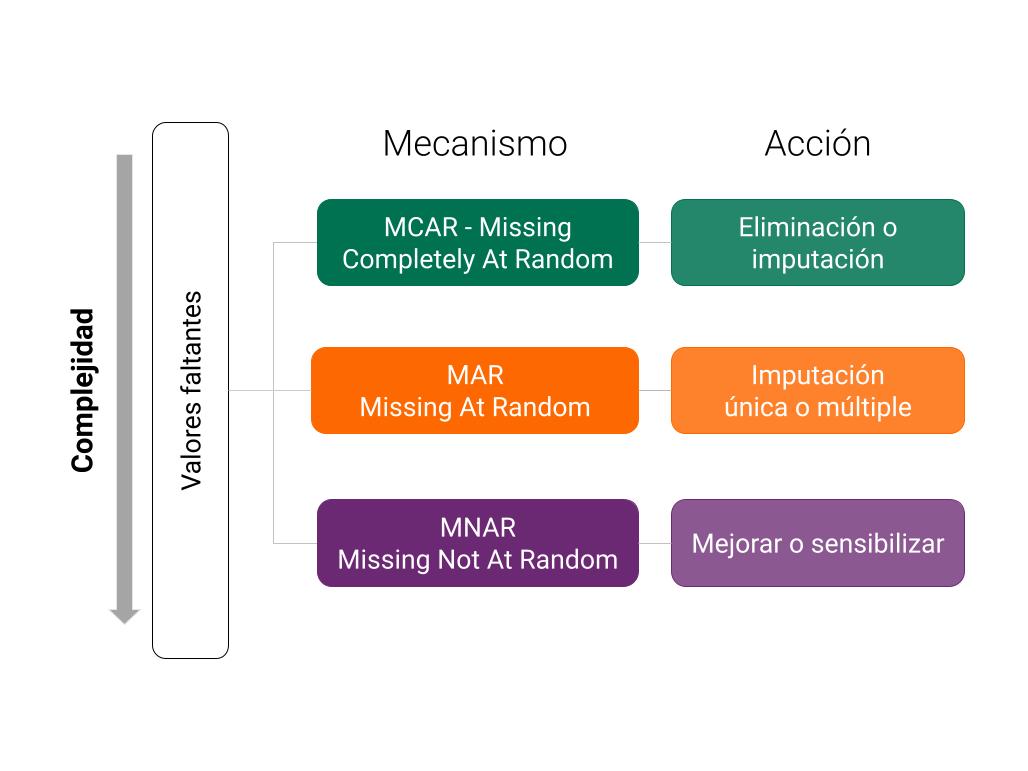
Manejo de Datos Faltantes: Imputación#
Configuración de ambiente de trabajo#
pip install -r requirements.txt
^C
Requirement already satisfied: pip in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 1)) (23.1.2)
Collecting pyjanitor (from -r requirements.txt (line 2))
Using cached pyjanitor-0.25.0-py3-none-any.whl (171 kB)
Requirement already satisfied: matplotlib in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 3)) (3.7.2)
Collecting missingno (from -r requirements.txt (line 4))
Using cached missingno-0.5.2-py3-none-any.whl (8.7 kB)
Requirement already satisfied: numpy in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 5)) (1.25.2)
Requirement already satisfied: pandas in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 6)) (2.1.0)
Requirement already satisfied: pandas-flavor in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 7)) (0.6.0)
Collecting pandas-profiling (from -r requirements.txt (line 8))
Using cached pandas_profiling-3.2.0-py2.py3-none-any.whl (262 kB)
Requirement already satisfied: seaborn in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 9)) (0.12.2)
Collecting session-info (from -r requirements.txt (line 10))
Using cached session_info-1.0.0.tar.gz (24 kB)
Preparing metadata (setup.py): started
Preparing metadata (setup.py): finished with status 'done'
Collecting ipywidgets (from -r requirements.txt (line 11))
Using cached ipywidgets-8.1.0-py3-none-any.whl (139 kB)
Collecting ozon3 (from -r requirements.txt (line 12))
Using cached ozon3-4.0.2-py3-none-any.whl (27 kB)
Collecting pyreadr (from -r requirements.txt (line 13))
Using cached pyreadr-0.4.9-cp311-cp311-win_amd64.whl (1.3 MB)
Collecting upsetplot (from -r requirements.txt (line 14))
Using cached UpSetPlot-0.8.0.tar.gz (22 kB)
Preparing metadata (setup.py): started
Preparing metadata (setup.py): finished with status 'done'
Collecting statsmodels==0.13.2 (from -r requirements.txt (line 15))
Using cached statsmodels-0.13.2.tar.gz (17.9 MB)
Installing build dependencies: started
Note: you may need to restart the kernel to use updated packages.
Importar librerías#
import janitor
import matplotlib.pyplot as plt
import missingno
import nhanes.load
import numpy as np
import pandas as pd
import scipy.stats
import seaborn as sns
import session_info
import sklearn.compose
import sklearn.impute
import sklearn.preprocessing
import statsmodels.api as sm
import statsmodels.datasets
import statsmodels.formula.api as smf
from sklearn.ensemble import RandomForestRegressor
from sklearn.experimental import enable_iterative_imputer
from sklearn.kernel_approximation import Nystroem
from sklearn.linear_model import BayesianRidge, Ridge
from sklearn.neighbors import KNeighborsRegressor
from statsmodels.graphics.mosaicplot import mosaic
Importar funciones personalizadas#
%run pandas-missing-extension.ipynb
/root/venv/lib/python3.9/site-packages/upsetplot/plotting.py:20: MatplotlibDeprecationWarning: The matplotlib.tight_layout module was deprecated in Matplotlib 3.6 and will be removed two minor releases later.
from matplotlib.tight_layout import get_renderer
Configurar el aspecto general de las gráficas del proyecto#
%matplotlib inline
sns.set(
rc={
"figure.figsize": (8, 6)
}
)
sns.set_style("whitegrid")
El problema de trabajar con valores faltantes#
airquality_df = (
sm.datasets.get_rdataset("airquality")
.data
.clean_names(
case_type = "snake"
)
.add_column("year", 1973)
.assign(
date = lambda df: pd.to_datetime(df[["year", "month", "day"]])
)
.sort_values(by = "date")
.set_index("date")
)
airquality_df
| ozone | solar_r | wind | temp | month | day | year | |
|---|---|---|---|---|---|---|---|
| date | |||||||
| 1973-05-01 | 41.0 | 190.0 | 7.4 | 67 | 5 | 1 | 1973 |
| 1973-05-02 | 36.0 | 118.0 | 8.0 | 72 | 5 | 2 | 1973 |
| 1973-05-03 | 12.0 | 149.0 | 12.6 | 74 | 5 | 3 | 1973 |
| 1973-05-04 | 18.0 | 313.0 | 11.5 | 62 | 5 | 4 | 1973 |
| 1973-05-05 | NaN | NaN | 14.3 | 56 | 5 | 5 | 1973 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 1973-09-26 | 30.0 | 193.0 | 6.9 | 70 | 9 | 26 | 1973 |
| 1973-09-27 | NaN | 145.0 | 13.2 | 77 | 9 | 27 | 1973 |
| 1973-09-28 | 14.0 | 191.0 | 14.3 | 75 | 9 | 28 | 1973 |
| 1973-09-29 | 18.0 | 131.0 | 8.0 | 76 | 9 | 29 | 1973 |
| 1973-09-30 | 20.0 | 223.0 | 11.5 | 68 | 9 | 30 | 1973 |
153 rows × 7 columns
Hagamos dos modelos de regresion para predecir la temperatura en funcion de otras variables:
(
smf.ols(
formula = "temp ~ ozone",
data = airquality_df
)
.fit()
.summary()
.tables[0]
)
| Dep. Variable: | temp | R-squared: | 0.488 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.483 |
| Method: | Least Squares | F-statistic: | 108.5 |
| Date: | Wed, 25 Jan 2023 | Prob (F-statistic): | 2.93e-18 |
| Time: | 03:02:38 | Log-Likelihood: | -386.27 |
| No. Observations: | 116 | AIC: | 776.5 |
| Df Residuals: | 114 | BIC: | 782.1 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
(
smf.ols(
formula = "temp ~ ozone + solar_r",
data = airquality_df
)
.fit()
.summary()
.tables[0]
)
| Dep. Variable: | temp | R-squared: | 0.491 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.481 |
| Method: | Least Squares | F-statistic: | 52.07 |
| Date: | Wed, 25 Jan 2023 | Prob (F-statistic): | 1.47e-16 |
| Time: | 03:02:38 | Log-Likelihood: | -369.78 |
| No. Observations: | 111 | AIC: | 745.6 |
| Df Residuals: | 108 | BIC: | 753.7 |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
Si analizas el numero de observaciones, veras que un modelo ignora cierta cantidad de valores faltantes mientras que el otro no lo hace. No es recomendable en este caso usar el R^2 para comparar ambos modelos como equivalentes. Por lo tanto toca imputar datos y asi poder realizarlo
Preparando datos: National Health and Nutrition Examination Survey#
%run download-data-and-load-it.ipynb
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:73: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax0.grid(b=False)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:142: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax1.grid(b=False)
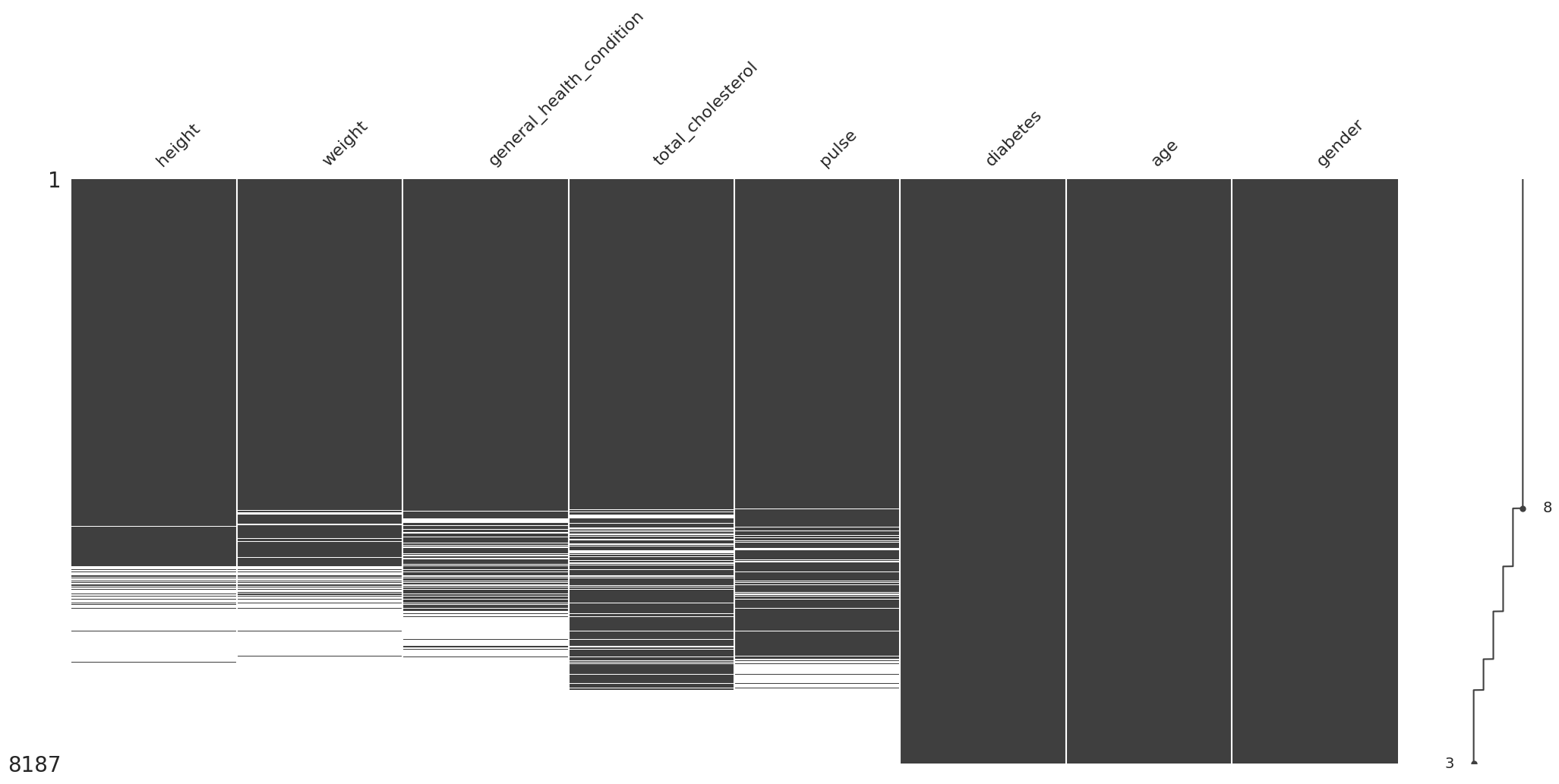
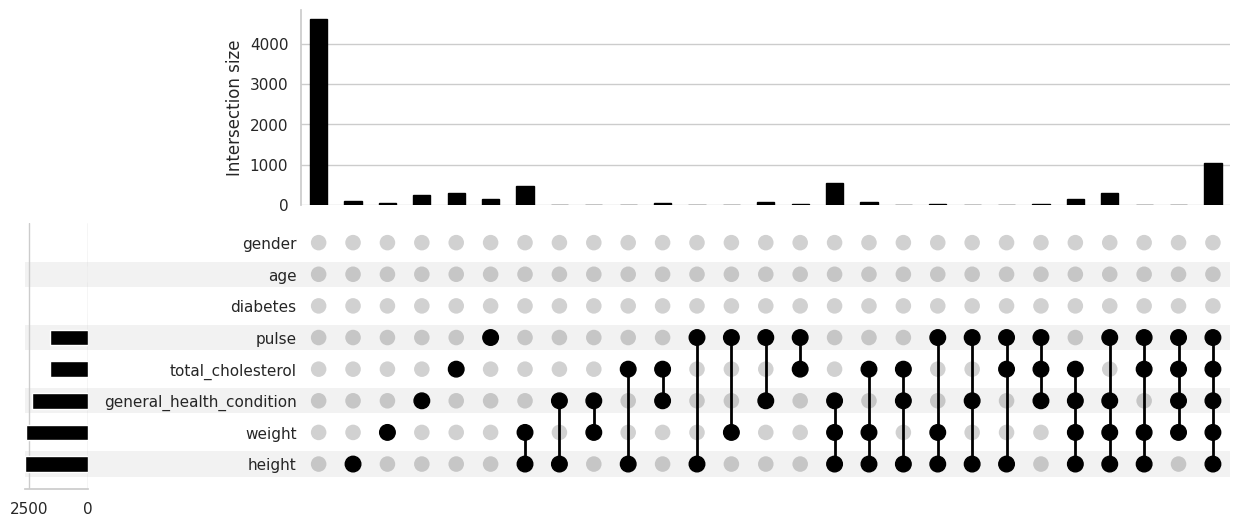
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:73: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax0.grid(b=False)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:142: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax1.grid(b=False)
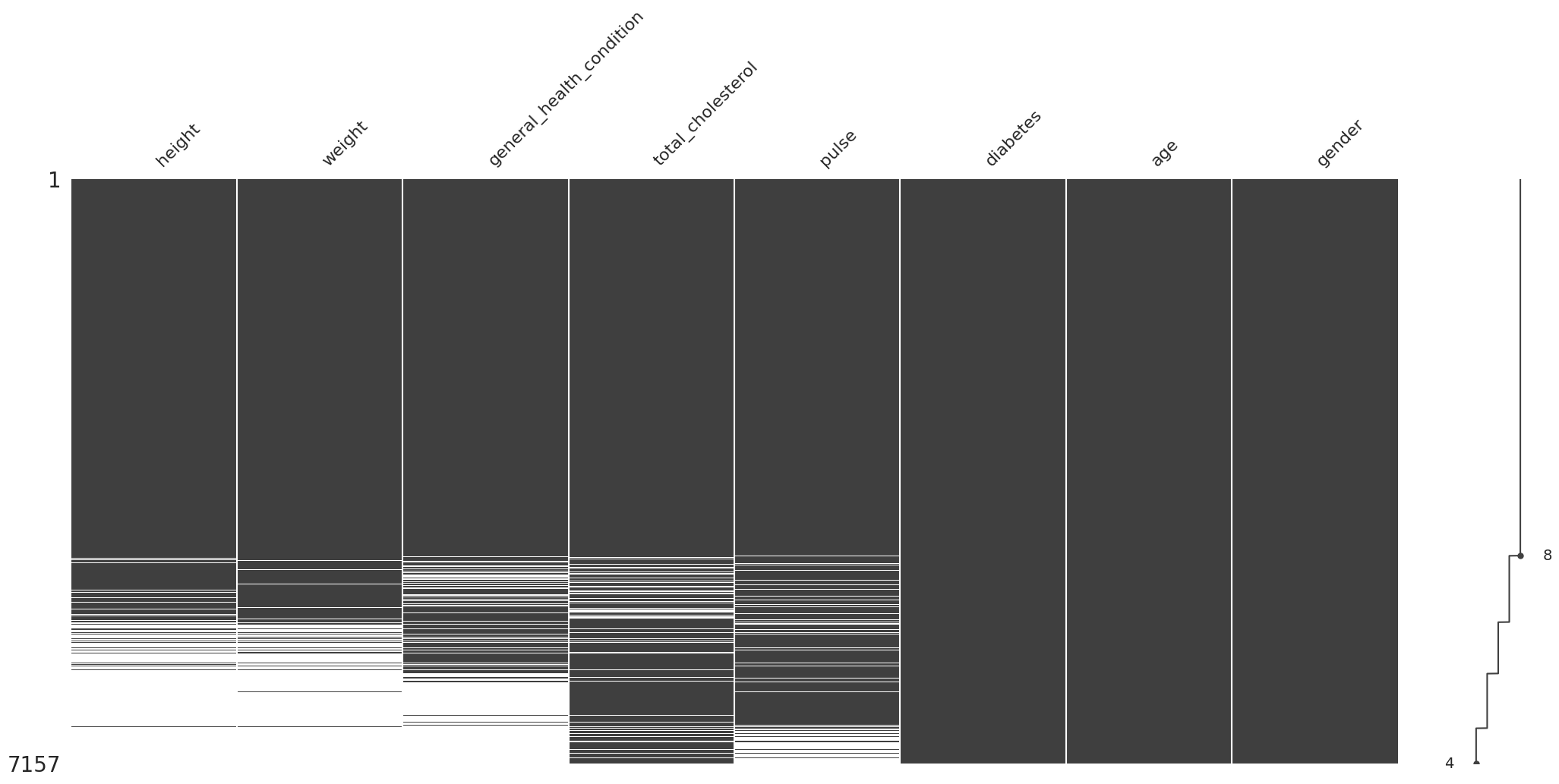
Consideración y evaluación de los distintos tipos de valores faltantes#
Evaluación del mecanismo de valores faltantes por prueba de t-test#
two-sided: las medias de las distribuciones subyacentes a las muestras son desiguales.
less: la media de la distribución subyacente a la primera muestra es menor que la media de la distribución subyacente a la segunda muestra.
greater: la media de la distribución subyacente a la primera muestra es mayor que la media de la distribución subyacente a la segunda muestra.
female_weight, male_weight = (
nhanes_df
.select_columns("gender", "weight")
.transform_column(
"weight",
lambda x: x.isna(),
elementwise = False
)
.groupby("gender")
.weight
.pipe(
lambda df: (
df.get_group("Female"),
df.get_group("Male")
)
)
)
# Ahora se hace una prueba estadistica para determinar si hay o no diferencia en la presencia
# o ausencia de valores de peso
scipy.stats.ttest_ind(
a = female_weight,
b = male_weight,
alternative="two-sided"
)
Ttest_indResult(statistic=-0.3621032192538131, pvalue=0.7172855918077239)
Dado que pvalue < 0.05, no se rechaza la hipotesis nula.
No existe diferencia estadistica entre los valores faltantes
Los datos entonces no estan perdidos al azar entre hombres y mujeres
Tratamiento de variables categóricas para imputación de valores faltantes#
# Primero se realiza una copia del dataframe original
nhanes_transformed_df = nhanes_df.copy(deep=True)
Codificación ordinal#
Una codificación ordinal implica mapear cada etiqueta (categoría) única a un valor entero. A su vez, la codificación ordinal también es conocida como codificación entera.
Ejemplo#
Dado un conjunto de datos con dos características, encontraremos los valores únicos por cataracterística y los transformaremos utilizando una codificación ordinal.
encoder = sklearn.preprocessing.OrdinalEncoder()
X = [["Male"], ["Female"], ["Female"]]
X
[['Male'], ['Female'], ['Female']]
encoder.fit_transform(X)
array([[1.],
[0.],
[0.]])
encoder.categories_
[array(['Female', 'Male'], dtype=object)]
encoder.inverse_transform([[1], [0], [0]])
array([['Male'],
['Female'],
['Female']], dtype=object)
Aplicando la codificación ordinal a todas tus variables categóricas#
categorical_columns = nhanes_df.select_dtypes(include=[object, "category"]).columns
categorical_transformer = sklearn.compose.make_column_transformer(
(sklearn.preprocessing.OrdinalEncoder(), categorical_columns),
remainder="passthrough"
#aqui le dices que no modifique las variables que no son categoricas
)
nhanes_transformed_df = (
pd.DataFrame(
categorical_transformer.fit_transform(nhanes_df),
columns = categorical_transformer.get_feature_names_out(),
index = nhanes_df.index
)
.rename_columns(
function = lambda x: x.removeprefix("ordinalencoder__")
)
.rename_columns(
function = lambda x: x.removeprefix("remainder__")
)
)
nhanes_transformed_df
| general_health_condition | gender | height | weight | total_cholesterol | pulse | diabetes | age | |
|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||
| 93705.0 | 2.0 | 0.0 | 63.0 | 165.0 | 157.0 | 52.0 | 0.0 | 66.0 |
| 93706.0 | 4.0 | 1.0 | 68.0 | 145.0 | 148.0 | 82.0 | 0.0 | 18.0 |
| 93707.0 | 2.0 | 1.0 | NaN | NaN | 189.0 | 100.0 | 0.0 | 13.0 |
| 93709.0 | NaN | 0.0 | 62.0 | 200.0 | 176.0 | 74.0 | 0.0 | 75.0 |
| 93711.0 | 4.0 | 1.0 | 69.0 | 142.0 | 238.0 | 62.0 | 0.0 | 56.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | 0.0 | 1.0 | 72.0 | 180.0 | 201.0 | 96.0 | 0.0 | 33.0 |
| 102953.0 | 1.0 | 1.0 | 65.0 | 218.0 | 182.0 | 78.0 | 0.0 | 42.0 |
| 102954.0 | 2.0 | 0.0 | 66.0 | 150.0 | 172.0 | 78.0 | 0.0 | 41.0 |
| 102955.0 | 4.0 | 0.0 | NaN | NaN | 150.0 | 74.0 | 0.0 | 14.0 |
| 102956.0 | 2.0 | 1.0 | 69.0 | 250.0 | 163.0 | 76.0 | 0.0 | 38.0 |
7157 rows × 8 columns
One Hot Encoding#
nhanes_transformed_df2 = nhanes_df.copy(deep=True)
pandas.get_dummies() vs skelearn.preprocessing.OneHotEncoder()#
pandas.get_dummies()#
pandas.get_dummies() tiene desventajas comparado con sklearn.preprocessing.onehotencoder(), estas son:
No sabe lidiar con valores nulos
Si no se seleccionan todas las columnas, puede que no se detecten todas las categorias
(
nhanes_transformed_df2
.select_columns("general_health_condition")
.pipe(pd.get_dummies)
)
| general_health_condition_Excellent | general_health_condition_Fair or | general_health_condition_Good | general_health_condition_Poor? | general_health_condition_Very good | |
|---|---|---|---|---|---|
| SEQN | |||||
| 93705.0 | 0 | 0 | 1 | 0 | 0 |
| 93706.0 | 0 | 0 | 0 | 0 | 1 |
| 93707.0 | 0 | 0 | 1 | 0 | 0 |
| 93709.0 | 0 | 0 | 0 | 0 | 0 |
| 93711.0 | 0 | 0 | 0 | 0 | 1 |
| ... | ... | ... | ... | ... | ... |
| 102949.0 | 1 | 0 | 0 | 0 | 0 |
| 102953.0 | 0 | 1 | 0 | 0 | 0 |
| 102954.0 | 0 | 0 | 1 | 0 | 0 |
| 102955.0 | 0 | 0 | 0 | 0 | 1 |
| 102956.0 | 0 | 0 | 1 | 0 | 0 |
7157 rows × 5 columns
skelearn.preprocessing.OneHotEncoder()#
Esta opcion tiene la ventaja que va a detectar automaticamente todas las categorias
transformer = sklearn.compose.make_column_transformer(
(sklearn.preprocessing.OrdinalEncoder(), ["gender"]),
(sklearn.preprocessing.OneHotEncoder(), ["general_health_condition"]),
remainder="passthrough"
)
nhanes_transformed_df2 = (
pd.DataFrame(
transformer.fit_transform(nhanes_df),
columns = transformer.get_feature_names_out(),
index = nhanes_df.index
)
.rename_columns(
function = lambda x: x.removeprefix("ordinalencoder__")
)
.rename_columns(
function = lambda x: x.removeprefix("remainder__")
)
.rename_columns(
function = lambda x: x.removeprefix("onehotencoder__")
)
)
nhanes_transformed_df2
| gender | general_health_condition_Excellent | general_health_condition_Fair or | general_health_condition_Good | general_health_condition_Poor? | general_health_condition_Very good | general_health_condition_nan | height | weight | total_cholesterol | pulse | diabetes | age | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SEQN | |||||||||||||
| 93705.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 63.0 | 165.0 | 157.0 | 52.0 | 0.0 | 66.0 |
| 93706.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 68.0 | 145.0 | 148.0 | 82.0 | 0.0 | 18.0 |
| 93707.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | NaN | NaN | 189.0 | 100.0 | 0.0 | 13.0 |
| 93709.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 62.0 | 200.0 | 176.0 | 74.0 | 0.0 | 75.0 |
| 93711.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 69.0 | 142.0 | 238.0 | 62.0 | 0.0 | 56.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 72.0 | 180.0 | 201.0 | 96.0 | 0.0 | 33.0 |
| 102953.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 65.0 | 218.0 | 182.0 | 78.0 | 0.0 | 42.0 |
| 102954.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 66.0 | 150.0 | 172.0 | 78.0 | 0.0 | 41.0 |
| 102955.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | NaN | NaN | 150.0 | 74.0 | 0.0 | 14.0 |
| 102956.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 69.0 | 250.0 | 163.0 | 76.0 | 0.0 | 38.0 |
7157 rows × 13 columns
(
transformer
.named_transformers_
.get("onehotencoder")
.categories_
)
[array(['Excellent', 'Fair or', 'Good', 'Poor?', 'Very good', nan],
dtype=object)]
(
transformer
.named_transformers_
.get("onehotencoder")
.inverse_transform(
X = [[0, 0, 1, 0, 0, 0]]
)
)
array([['Good']], dtype=object)
Tipos de imputación de valores faltantes#
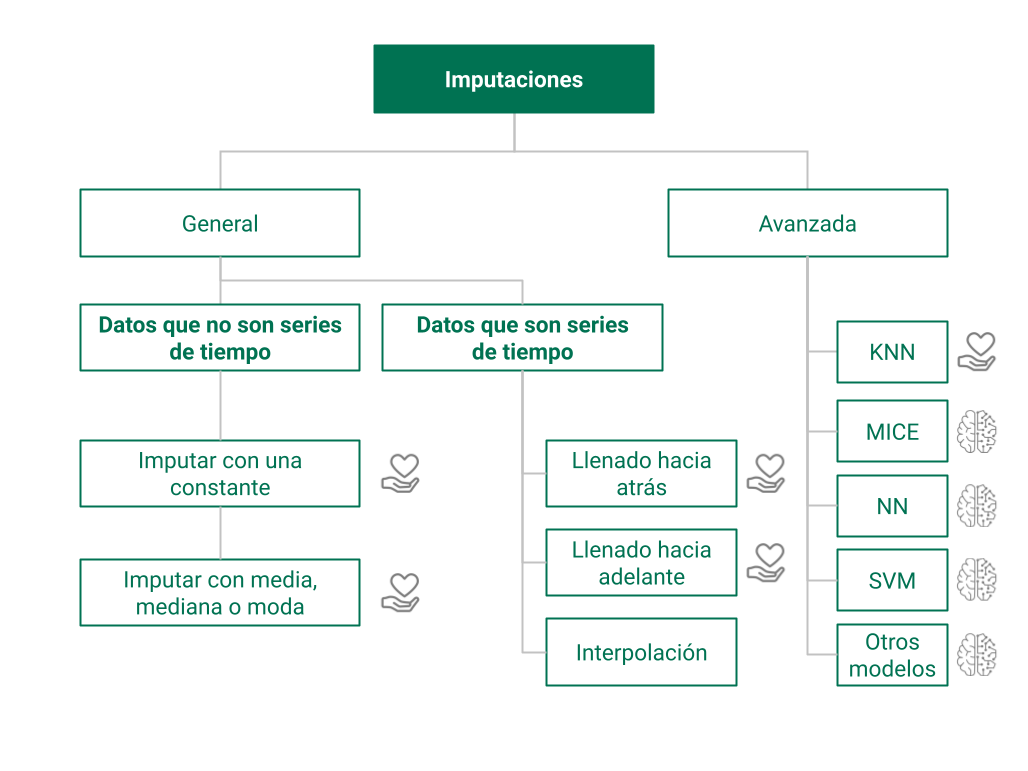
Imputación de un único valor (media, mediana, moda)#
Media#
(
nhanes_df
# janitor
.transform_column(
"height",
lambda x: x.fillna(x.mean()),
elementwise=False
)
)
| height | weight | general_health_condition | total_cholesterol | pulse | diabetes | age | gender | |
|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||
| 93705.0 | 63.00000 | 165.0 | Good | 157.0 | 52.0 | 0 | 66.0 | Female |
| 93706.0 | 68.00000 | 145.0 | Very good | 148.0 | 82.0 | 0 | 18.0 | Male |
| 93707.0 | 66.25656 | NaN | Good | 189.0 | 100.0 | 0 | 13.0 | Male |
| 93709.0 | 62.00000 | 200.0 | NaN | 176.0 | 74.0 | 0 | 75.0 | Female |
| 93711.0 | 69.00000 | 142.0 | Very good | 238.0 | 62.0 | 0 | 56.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | 72.00000 | 180.0 | Excellent | 201.0 | 96.0 | 0 | 33.0 | Male |
| 102953.0 | 65.00000 | 218.0 | Fair or | 182.0 | 78.0 | 0 | 42.0 | Male |
| 102954.0 | 66.00000 | 150.0 | Good | 172.0 | 78.0 | 0 | 41.0 | Female |
| 102955.0 | 66.25656 | NaN | Very good | 150.0 | 74.0 | 0 | 14.0 | Female |
| 102956.0 | 69.00000 | 250.0 | Good | 163.0 | 76.0 | 0 | 38.0 | Male |
7157 rows × 8 columns
(
nhanes_df
.select_columns("height", "weight")
.missing.bind_shadow_matrix(True,False, suffix = "_imp")
.assign(
height = lambda df: df.height.fillna(value = df.height.mean()),
weight = lambda df: df.weight.fillna(value = df.weight.mean())
)
.missing.scatter_imputation_plot(
x = "height",
y = "weight"
)
)
<AxesSubplot: xlabel='height', ylabel='weight'>
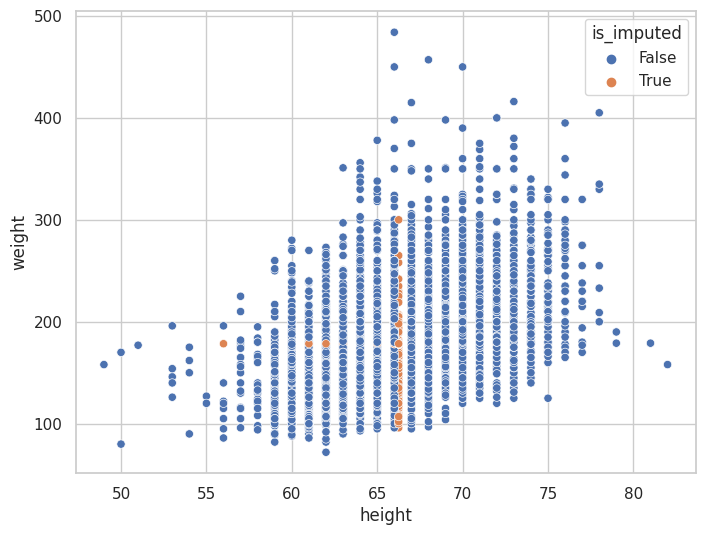
(
nhanes_df
.select_columns("height", "weight")
.missing.bind_shadow_matrix(True,False, suffix = "_imp")
.assign(
height = lambda df: df.height.fillna(value = df.height.mean()),
weight = lambda df: df.weight.fillna(value = df.weight.mean())
)
.missing.scatter_imputation_plot(
x = "height",
y = "weight",
show_marginal= True,
height = 10
)
)
<seaborn.axisgrid.JointGrid at 0x7f756fdf28b0>
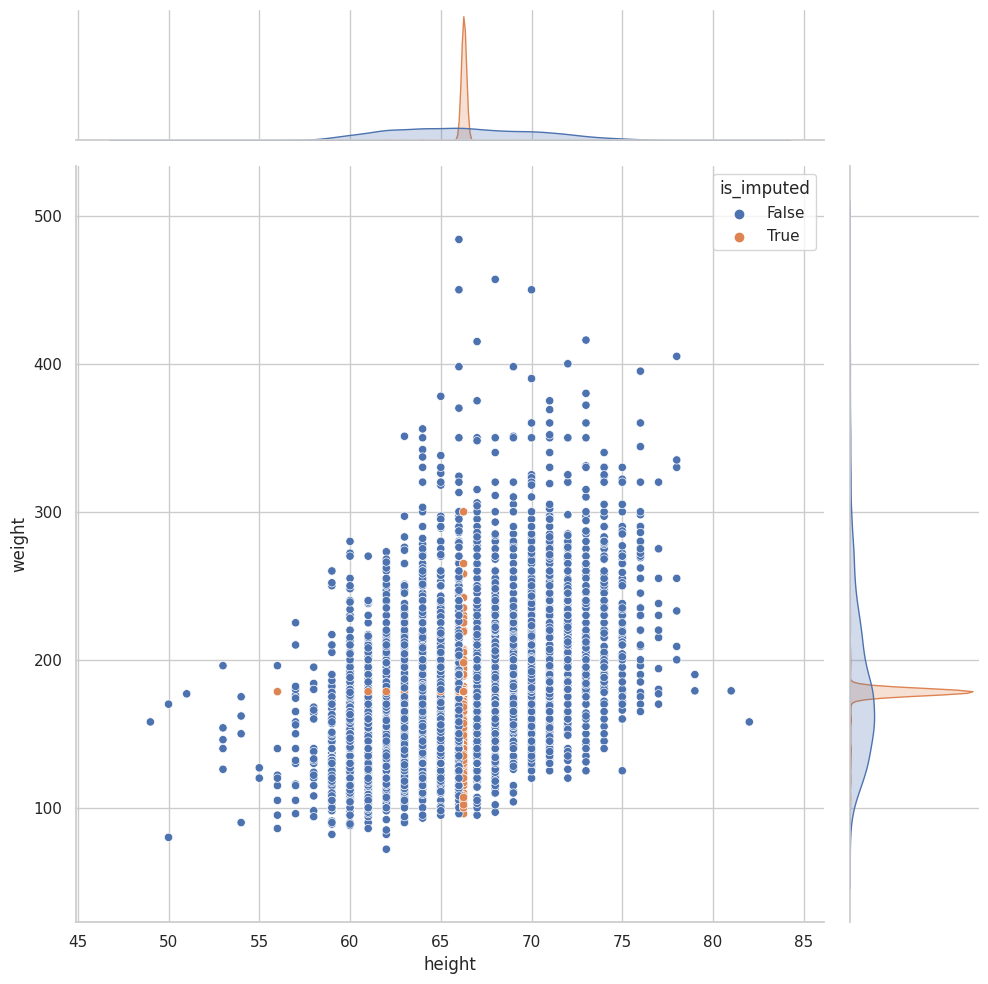
Imputación por llenado hacia atrás e imputación por llenado hacia adelante#
fillna() vs ffill() o bfill()#
Forwardfill#
Imputa el dato de la fila anterior en nuestro missing value
(
nhanes_df
.select_columns("height", "weight")
# .fillna(method = "ffill")
.ffill()
)
| height | weight | |
|---|---|---|
| SEQN | ||
| 93705.0 | 63.0 | 165.0 |
| 93706.0 | 68.0 | 145.0 |
| 93707.0 | 68.0 | 145.0 |
| 93709.0 | 62.0 | 200.0 |
| 93711.0 | 69.0 | 142.0 |
| ... | ... | ... |
| 102949.0 | 72.0 | 180.0 |
| 102953.0 | 65.0 | 218.0 |
| 102954.0 | 66.0 | 150.0 |
| 102955.0 | 66.0 | 150.0 |
| 102956.0 | 69.0 | 250.0 |
7157 rows × 2 columns
Backfill#
Imputa el dato de la fila posterior en nuestro missing value
(
nhanes_df
.select_columns("height", "weight")
# .fillna(method = "bfill")
.bfill()
)
| height | weight | |
|---|---|---|
| SEQN | ||
| 93705.0 | 63.0 | 165.0 |
| 93706.0 | 68.0 | 145.0 |
| 93707.0 | 62.0 | 200.0 |
| 93709.0 | 62.0 | 200.0 |
| 93711.0 | 69.0 | 142.0 |
| ... | ... | ... |
| 102949.0 | 72.0 | 180.0 |
| 102953.0 | 65.0 | 218.0 |
| 102954.0 | 66.0 | 150.0 |
| 102955.0 | 69.0 | 250.0 |
| 102956.0 | 69.0 | 250.0 |
7157 rows × 2 columns
Recomendaciones al imputar valores utilizando ffill() o bfill()#
Imputación dentro de dominios e imputación a través de variables correlacionadas
Se recomienda organizar por grupos similares para que no se imputen datos aleatoriamente, sino que el dato que se va a imputar tenga sentido
(
nhanes_df
.select_columns("height", "weight", "gender", "diabetes", "general_health_condition")
.sort_values(
by = ["gender", "diabetes", "general_health_condition", "height"],
ascending = True
)
.transform_column(
"weight",
lambda x: x.ffill(),
elementwise = False
)
)
| height | weight | gender | diabetes | general_health_condition | |
|---|---|---|---|---|---|
| SEQN | |||||
| 94421.0 | 56.0 | 115.0 | Female | 0 | Excellent |
| 94187.0 | 59.0 | 130.0 | Female | 0 | Excellent |
| 95289.0 | 59.0 | 162.0 | Female | 0 | Excellent |
| 97967.0 | 59.0 | 130.0 | Female | 0 | Excellent |
| 99125.0 | 59.0 | 105.0 | Female | 0 | Excellent |
| ... | ... | ... | ... | ... | ... |
| 96561.0 | 74.0 | 290.0 | Male | 1 | NaN |
| 96954.0 | NaN | 175.0 | Male | 1 | NaN |
| 97267.0 | NaN | 175.0 | Male | 1 | NaN |
| 97856.0 | NaN | 175.0 | Male | 1 | NaN |
| 98317.0 | NaN | 175.0 | Male | 1 | NaN |
7157 rows × 5 columns
(
nhanes_df
.select_columns("height", "weight", "gender", "diabetes", "general_health_condition")
.sort_values(
by=['gender','diabetes','general_health_condition','weight'],
ascending=True
)
.groupby(["gender", "general_health_condition"], dropna=False)
.apply(lambda x: x.ffill())
)
/tmp/ipykernel_1384/909901090.py:2: FutureWarning: Not prepending group keys to the result index of transform-like apply. In the future, the group keys will be included in the index, regardless of whether the applied function returns a like-indexed object.
To preserve the previous behavior, use
>>> .groupby(..., group_keys=False)
To adopt the future behavior and silence this warning, use
>>> .groupby(..., group_keys=True)
nhanes_df
| height | weight | gender | diabetes | general_health_condition | |
|---|---|---|---|---|---|
| SEQN | |||||
| 94195.0 | 63.0 | 90.0 | Female | 0 | Excellent |
| 95793.0 | 61.0 | 96.0 | Female | 0 | Excellent |
| 101420.0 | 59.0 | 98.0 | Female | 0 | Excellent |
| 94148.0 | 65.0 | 100.0 | Female | 0 | Excellent |
| 102062.0 | 62.0 | 100.0 | Female | 0 | Excellent |
| ... | ... | ... | ... | ... | ... |
| 96561.0 | 74.0 | 290.0 | Male | 1 | NaN |
| 96869.0 | 72.0 | 298.0 | Male | 1 | NaN |
| 97267.0 | 72.0 | 298.0 | Male | 1 | NaN |
| 97856.0 | 72.0 | 298.0 | Male | 1 | NaN |
| 98317.0 | 72.0 | 298.0 | Male | 1 | NaN |
7157 rows × 5 columns
Imputación por interpolación#
(
airquality_df
.select_columns("ozone")
.pipe(
lambda df: (
df.ozone.plot(color = "#313638", marker= "o")
)
)
)
<AxesSubplot: xlabel='date'>
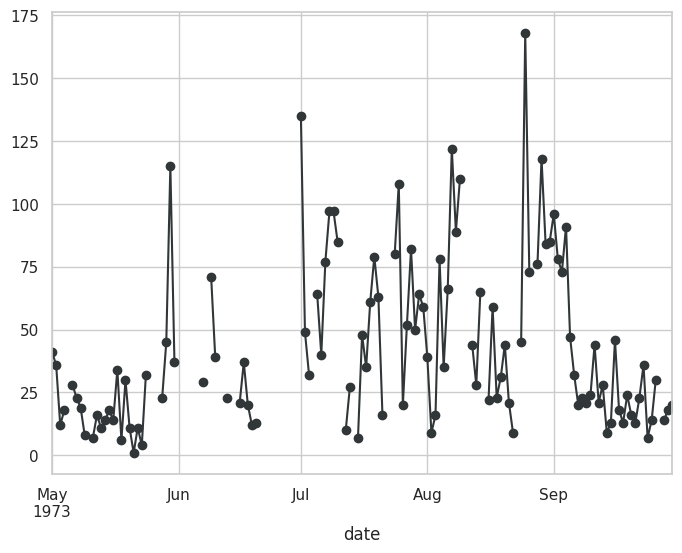
Visualizando con ffill#
plt.figure(figsize=(20,10))
(
airquality_df
.select_columns("ozone")
.pipe(
lambda df: (
df.ozone.ffill().plot(color= "red", marker="o", alpha=6/9, linestyle="dashed"),
df.ozone.plot(color = "#313638", marker= "o")
)
)
)
(<AxesSubplot: xlabel='date'>, <AxesSubplot: xlabel='date'>)
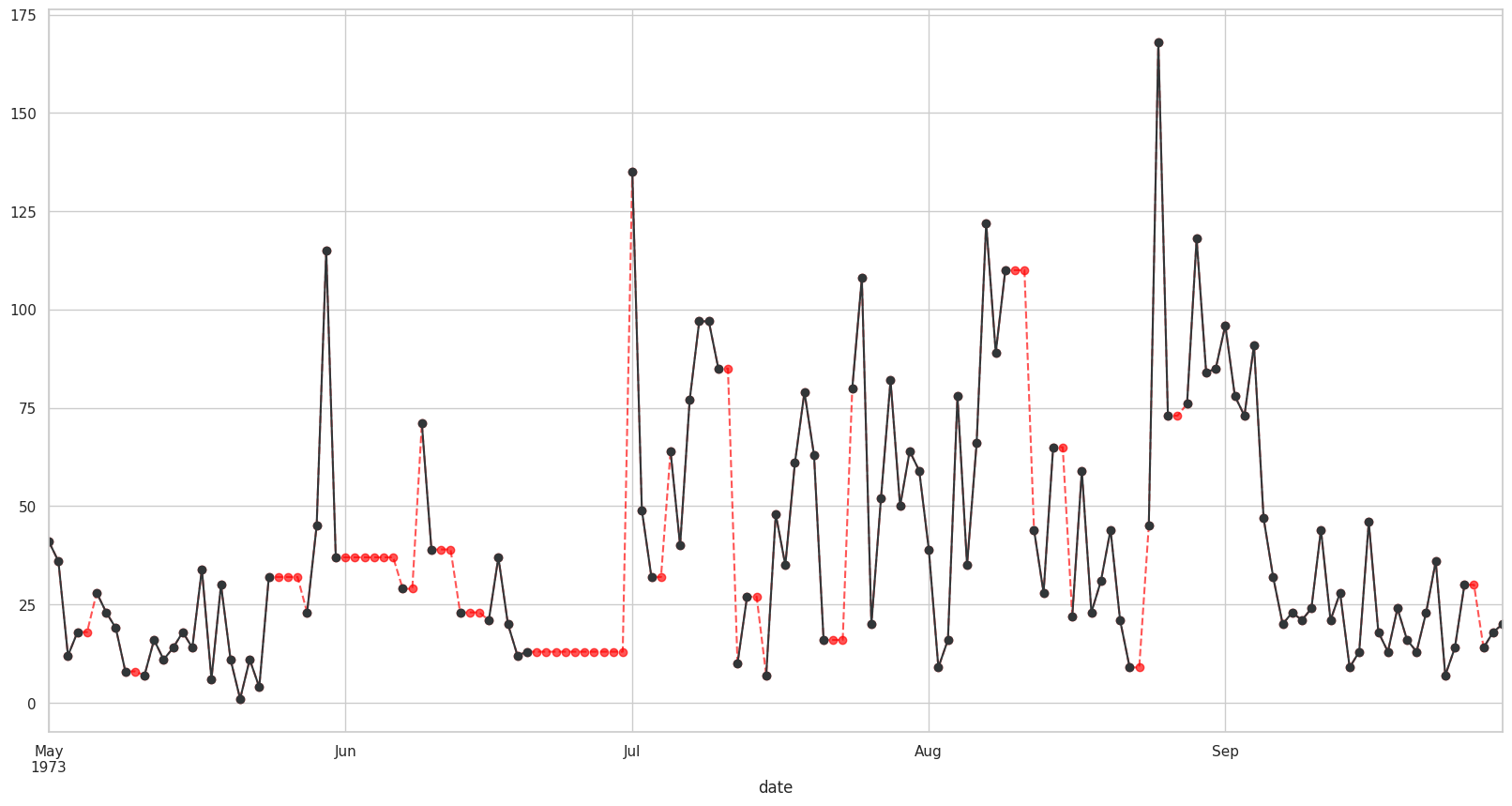
Visualizando con bfill#
plt.figure(figsize=(20,10))
(
airquality_df
.select_columns("ozone")
.pipe(
lambda df: (
df.ozone.bfill().plot(color= "red", marker="o", alpha=6/9, linestyle="dashed"),
df.ozone.plot(color = "#313638", marker= "o")
)
)
)
(<AxesSubplot: xlabel='date'>, <AxesSubplot: xlabel='date'>)
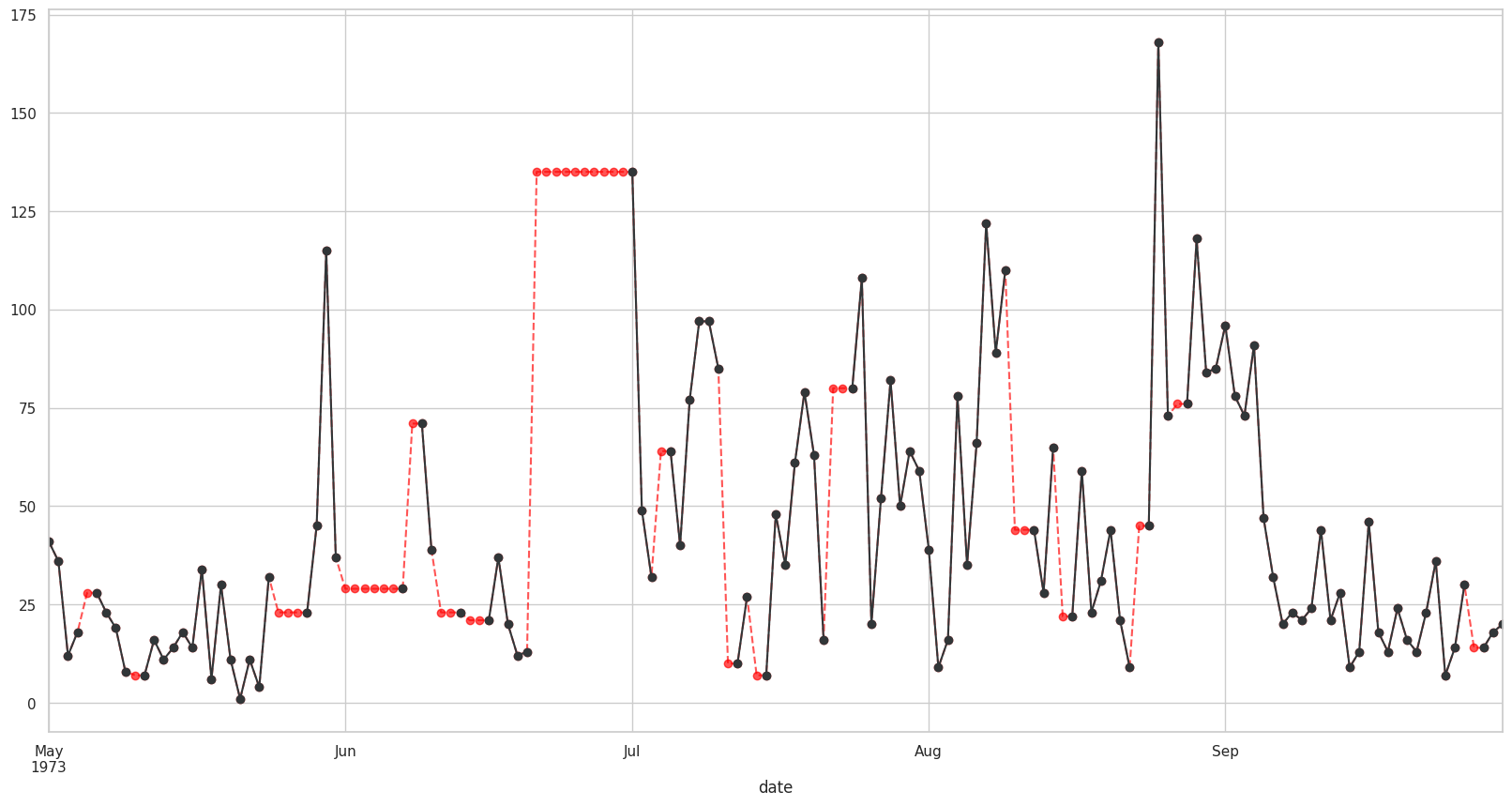
Visualizando con bfill o ffill con valores mas cercanos#
plt.figure(figsize=(20,10))
(
airquality_df
.select_columns("ozone")
.pipe(
lambda df: (
df.ozone.interpolate(method = "nearest").plot(color= "red", marker="o", alpha=6/9, linestyle="dashed"),
df.ozone.plot(color = "#313638", marker= "o")
)
)
)
(<AxesSubplot: xlabel='date'>, <AxesSubplot: xlabel='date'>)
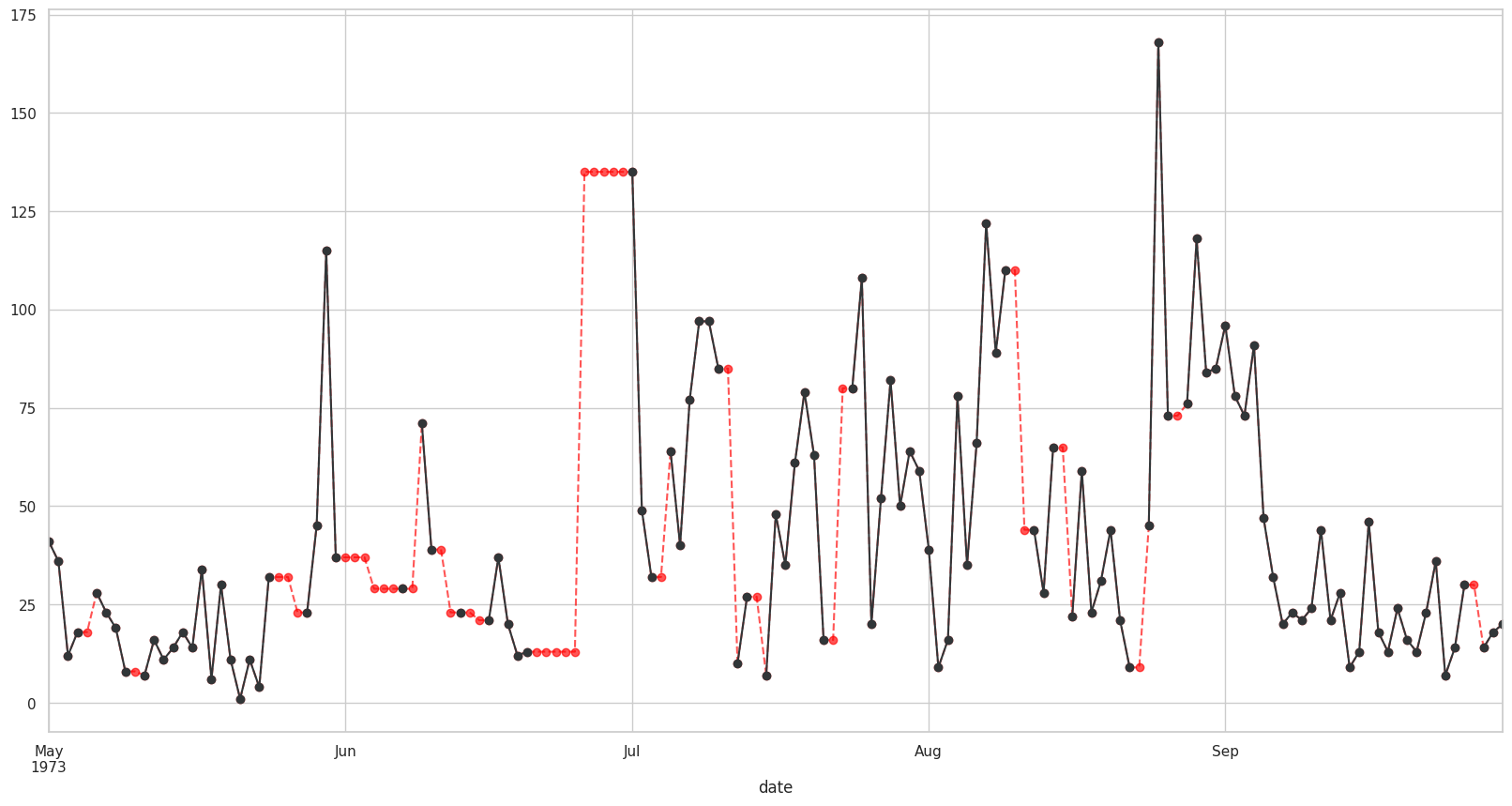
Visualizando valores interpolados#
plt.figure(figsize=(20,10))
(
airquality_df
.select_columns("ozone")
.pipe(
lambda df: (
df.ozone.interpolate(method = "linear").plot(color= "red", marker="o", alpha=6/9, linestyle="dashed"),
df.ozone.plot(color = "#313638", marker= "o")
)
)
)
(<AxesSubplot: xlabel='date'>, <AxesSubplot: xlabel='date'>)
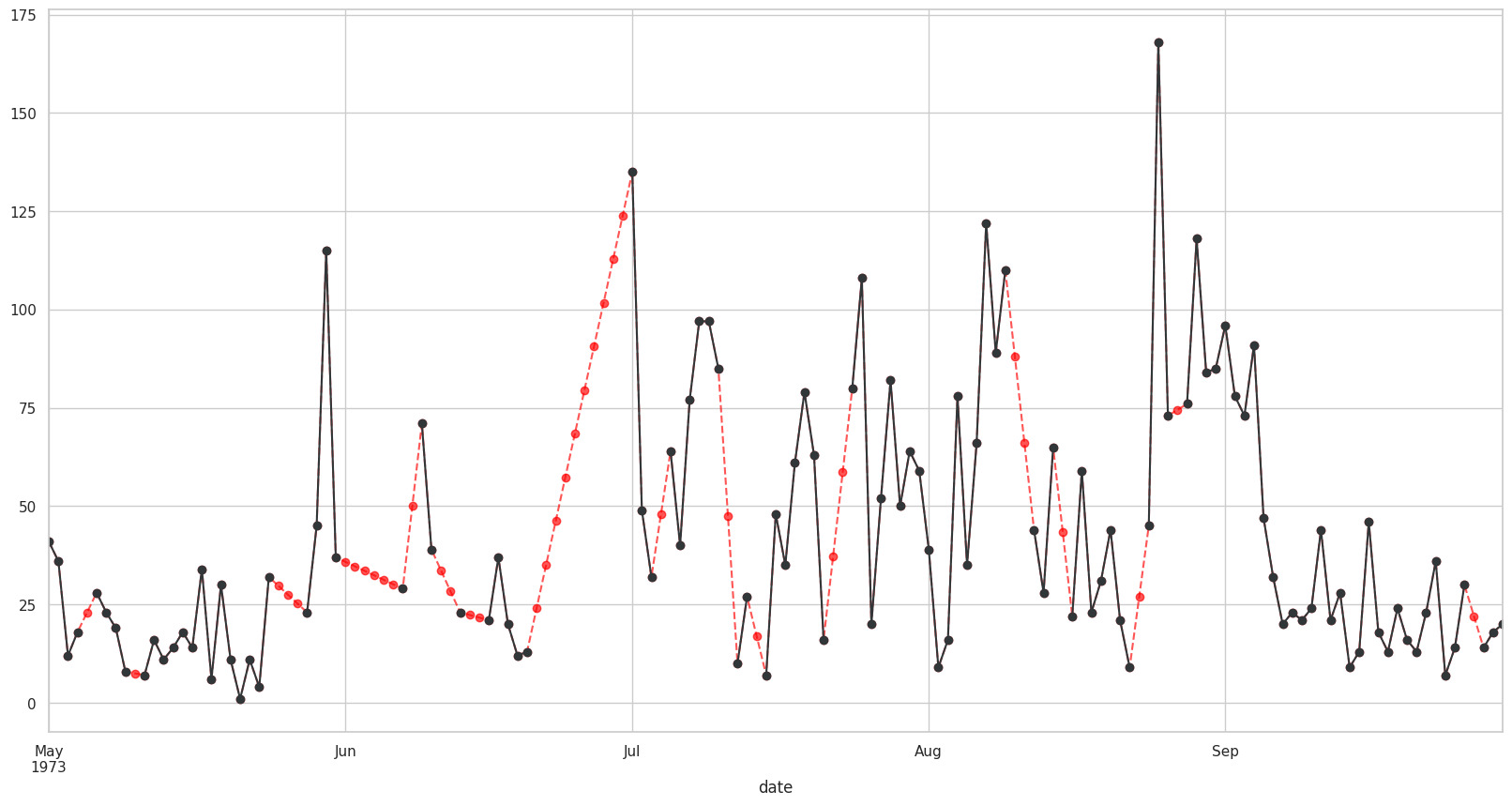
Ahora para asignar esos valores a la columna:
airquality_df["ozone"] = airquality_df.ozone.interpolate(method="linear")
airquality_df
| ozone | solar_r | wind | temp | month | day | year | |
|---|---|---|---|---|---|---|---|
| date | |||||||
| 1973-05-01 | 41.0 | 190.0 | 7.4 | 67 | 5 | 1 | 1973 |
| 1973-05-02 | 36.0 | 118.0 | 8.0 | 72 | 5 | 2 | 1973 |
| 1973-05-03 | 12.0 | 149.0 | 12.6 | 74 | 5 | 3 | 1973 |
| 1973-05-04 | 18.0 | 313.0 | 11.5 | 62 | 5 | 4 | 1973 |
| 1973-05-05 | 23.0 | NaN | 14.3 | 56 | 5 | 5 | 1973 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 1973-09-26 | 30.0 | 193.0 | 6.9 | 70 | 9 | 26 | 1973 |
| 1973-09-27 | 22.0 | 145.0 | 13.2 | 77 | 9 | 27 | 1973 |
| 1973-09-28 | 14.0 | 191.0 | 14.3 | 75 | 9 | 28 | 1973 |
| 1973-09-29 | 18.0 | 131.0 | 8.0 | 76 | 9 | 29 | 1973 |
| 1973-09-30 | 20.0 | 223.0 | 11.5 | 68 | 9 | 30 | 1973 |
153 rows × 7 columns
Imputación por algoritmo de vecinos más cercanos (KNN)#
K nearest neighbors (KNN) es el algritmo mas utilizado de imputacion
Resumen de métodos de distancia:
Euclidiana: Útil para variables numéricas
Manhattan: Útil paa variables tipo factor
Hamming: Útil para variables categóricas
Gower: Útil para conjuntos de datos con variables mixtas
nhanes_transformed_df
| general_health_condition | gender | height | weight | total_cholesterol | pulse | diabetes | age | |
|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||
| 93705.0 | 2.0 | 0.0 | 63.0 | 165.0 | 157.0 | 52.0 | 0.0 | 66.0 |
| 93706.0 | 4.0 | 1.0 | 68.0 | 145.0 | 148.0 | 82.0 | 0.0 | 18.0 |
| 93707.0 | 2.0 | 1.0 | NaN | NaN | 189.0 | 100.0 | 0.0 | 13.0 |
| 93709.0 | NaN | 0.0 | 62.0 | 200.0 | 176.0 | 74.0 | 0.0 | 75.0 |
| 93711.0 | 4.0 | 1.0 | 69.0 | 142.0 | 238.0 | 62.0 | 0.0 | 56.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | 0.0 | 1.0 | 72.0 | 180.0 | 201.0 | 96.0 | 0.0 | 33.0 |
| 102953.0 | 1.0 | 1.0 | 65.0 | 218.0 | 182.0 | 78.0 | 0.0 | 42.0 |
| 102954.0 | 2.0 | 0.0 | 66.0 | 150.0 | 172.0 | 78.0 | 0.0 | 41.0 |
| 102955.0 | 4.0 | 0.0 | NaN | NaN | 150.0 | 74.0 | 0.0 | 14.0 |
| 102956.0 | 2.0 | 1.0 | 69.0 | 250.0 | 163.0 | 76.0 | 0.0 | 38.0 |
7157 rows × 8 columns
KNN algorithm#
knn_imputer = sklearn.impute.KNNImputer()
#copia del df original
nhanes_df_knn = nhanes_transformed_df.copy(deep=True)
nhanes_df_knn.iloc[:, :] = knn_imputer.fit_transform(nhanes_transformed_df).round()
nhanes_df_knn
| general_health_condition | gender | height | weight | total_cholesterol | pulse | diabetes | age | |
|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||
| 93705.0 | 2.0 | 0.0 | 63.0 | 165.0 | 157.0 | 52.0 | 0.0 | 66.0 |
| 93706.0 | 4.0 | 1.0 | 68.0 | 145.0 | 148.0 | 82.0 | 0.0 | 18.0 |
| 93707.0 | 2.0 | 1.0 | 69.0 | 130.0 | 189.0 | 100.0 | 0.0 | 13.0 |
| 93709.0 | 2.0 | 0.0 | 62.0 | 200.0 | 176.0 | 74.0 | 0.0 | 75.0 |
| 93711.0 | 4.0 | 1.0 | 69.0 | 142.0 | 238.0 | 62.0 | 0.0 | 56.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | 0.0 | 1.0 | 72.0 | 180.0 | 201.0 | 96.0 | 0.0 | 33.0 |
| 102953.0 | 1.0 | 1.0 | 65.0 | 218.0 | 182.0 | 78.0 | 0.0 | 42.0 |
| 102954.0 | 2.0 | 0.0 | 66.0 | 150.0 | 172.0 | 78.0 | 0.0 | 41.0 |
| 102955.0 | 4.0 | 0.0 | 71.0 | 159.0 | 150.0 | 74.0 | 0.0 | 14.0 |
| 102956.0 | 2.0 | 1.0 | 69.0 | 250.0 | 163.0 | 76.0 | 0.0 | 38.0 |
7157 rows × 8 columns
(
pd.concat(
[
nhanes_df_knn,
nhanes_df.missing.create_shadow_matrix(True, False, suffix="_imp", only_missing=True)
],
axis=1
)
)
| general_health_condition | gender | height | weight | total_cholesterol | pulse | diabetes | age | height_imp | weight_imp | general_health_condition_imp | total_cholesterol_imp | pulse_imp | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SEQN | |||||||||||||
| 93705.0 | 2.0 | 0.0 | 63.0 | 165.0 | 157.0 | 52.0 | 0.0 | 66.0 | False | False | False | False | False |
| 93706.0 | 4.0 | 1.0 | 68.0 | 145.0 | 148.0 | 82.0 | 0.0 | 18.0 | False | False | False | False | False |
| 93707.0 | 2.0 | 1.0 | 69.0 | 130.0 | 189.0 | 100.0 | 0.0 | 13.0 | True | True | False | False | False |
| 93709.0 | 2.0 | 0.0 | 62.0 | 200.0 | 176.0 | 74.0 | 0.0 | 75.0 | False | False | True | False | False |
| 93711.0 | 4.0 | 1.0 | 69.0 | 142.0 | 238.0 | 62.0 | 0.0 | 56.0 | False | False | False | False | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | 0.0 | 1.0 | 72.0 | 180.0 | 201.0 | 96.0 | 0.0 | 33.0 | False | False | False | False | False |
| 102953.0 | 1.0 | 1.0 | 65.0 | 218.0 | 182.0 | 78.0 | 0.0 | 42.0 | False | False | False | False | False |
| 102954.0 | 2.0 | 0.0 | 66.0 | 150.0 | 172.0 | 78.0 | 0.0 | 41.0 | False | False | False | False | False |
| 102955.0 | 4.0 | 0.0 | 71.0 | 159.0 | 150.0 | 74.0 | 0.0 | 14.0 | True | True | False | False | False |
| 102956.0 | 2.0 | 1.0 | 69.0 | 250.0 | 163.0 | 76.0 | 0.0 | 38.0 | False | False | False | False | False |
7157 rows × 13 columns
(
pd.concat(
[
nhanes_df_knn,
nhanes_df.missing.create_shadow_matrix(True, False, suffix="_imp", only_missing=True)
],
axis=1
)
.missing.scatter_imputation_plot(
x = "height",
y = "weight"
)
)
<AxesSubplot: xlabel='height', ylabel='weight'>
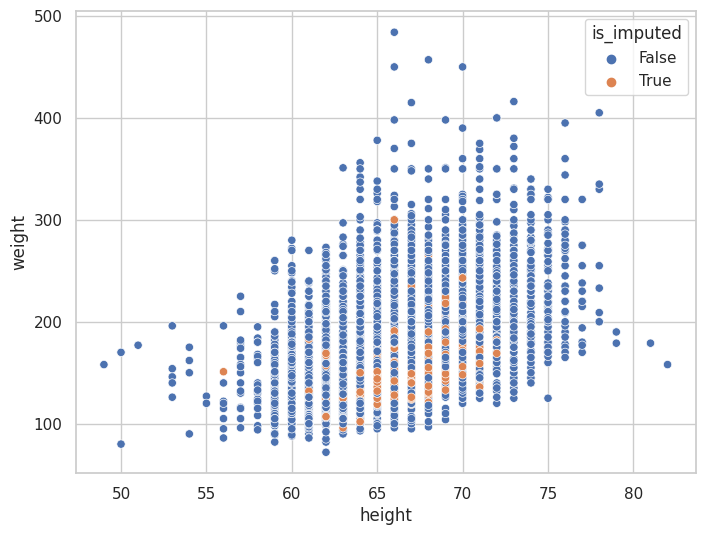
Ordenamiento por cantidad de variables faltantes#
knn_imputer = sklearn.impute.KNNImputer()
#copia del df original
nhanes_df_knn = nhanes_transformed_df.missing.sort_variables_by_missingness(ascending=True).copy(deep=True)
nhanes_df_knn.iloc[:, :] = knn_imputer.fit_transform(nhanes_transformed_df.missing.sort_variables_by_missingness(ascending=True)).round()
nhanes_df_knn
| gender | diabetes | age | pulse | total_cholesterol | general_health_condition | weight | height | |
|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||
| 93705.0 | 0.0 | 0.0 | 66.0 | 52.0 | 157.0 | 2.0 | 165.0 | 63.0 |
| 93706.0 | 1.0 | 0.0 | 18.0 | 82.0 | 148.0 | 4.0 | 145.0 | 68.0 |
| 93707.0 | 1.0 | 0.0 | 13.0 | 100.0 | 189.0 | 2.0 | 130.0 | 69.0 |
| 93709.0 | 0.0 | 0.0 | 75.0 | 74.0 | 176.0 | 2.0 | 200.0 | 62.0 |
| 93711.0 | 1.0 | 0.0 | 56.0 | 62.0 | 238.0 | 4.0 | 142.0 | 69.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | 1.0 | 0.0 | 33.0 | 96.0 | 201.0 | 0.0 | 180.0 | 72.0 |
| 102953.0 | 1.0 | 0.0 | 42.0 | 78.0 | 182.0 | 1.0 | 218.0 | 65.0 |
| 102954.0 | 0.0 | 0.0 | 41.0 | 78.0 | 172.0 | 2.0 | 150.0 | 66.0 |
| 102955.0 | 0.0 | 0.0 | 14.0 | 74.0 | 150.0 | 4.0 | 159.0 | 71.0 |
| 102956.0 | 1.0 | 0.0 | 38.0 | 76.0 | 163.0 | 2.0 | 250.0 | 69.0 |
7157 rows × 8 columns
(
pd.concat(
[
nhanes_df_knn,
nhanes_df.missing.create_shadow_matrix(True, False, suffix="_imp", only_missing=True)
],
axis=1
)
.missing.scatter_imputation_plot(
x = "height",
y = "weight"
)
)
<AxesSubplot: xlabel='height', ylabel='weight'>

Imputación basada en modelos#
nhanes_model_df = (
nhanes_df
.select_columns("height", "weight", "gender", "age")
.sort_values("weight")
.transform_column(
"weight",
lambda x: x.ffill(),
elementwise = False
)
.missing.bind_shadow_matrix(
True,
False,
suffix="_imp",
only_missing = False
)
)
nhanes_model_df
| height | weight | gender | age | height_imp | weight_imp | gender_imp | age_imp | |
|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||
| 99970.0 | 62.0 | 72.0 | Male | 16.0 | False | False | False | False |
| 97877.0 | 50.0 | 80.0 | Female | 29.0 | False | False | False | False |
| 96451.0 | 59.0 | 82.0 | Female | 80.0 | False | False | False | False |
| 97730.0 | 62.0 | 82.0 | Female | 19.0 | False | False | False | False |
| 99828.0 | 62.0 | 85.0 | Female | 16.0 | False | False | False | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102915.0 | NaN | 484.0 | Female | 14.0 | True | False | False | False |
| 102926.0 | NaN | 484.0 | Female | 15.0 | True | False | False | False |
| 102941.0 | NaN | 484.0 | Female | 14.0 | True | False | False | False |
| 102945.0 | NaN | 484.0 | Male | 15.0 | True | False | False | False |
| 102955.0 | NaN | 484.0 | Female | 14.0 | True | False | False | False |
7157 rows × 8 columns
height_ols = (
nhanes_model_df
.pipe(
lambda df: smf.ols("height ~ weight + age", data=df)
)
.fit()
)
ols_imputed_values = (
nhanes_model_df
.pipe(
lambda df: df[df.height.isna()]
)
.pipe(
lambda df: height_ols.predict(df).round()
)
)
ols_imputed_values
SEQN
100246.0 64.0
101209.0 64.0
102766.0 64.0
102509.0 64.0
96900.0 65.0
...
102915.0 75.0
102926.0 75.0
102941.0 75.0
102945.0 75.0
102955.0 75.0
Length: 1669, dtype: float64
nhanes_model_df.loc[nhanes_model_df.height.isna(), ["height"]] = ols_imputed_values
nhanes_model_df
| height | weight | gender | age | height_imp | weight_imp | gender_imp | age_imp | |
|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||
| 99970.0 | 62.0 | 72.0 | Male | 16.0 | False | False | False | False |
| 97877.0 | 50.0 | 80.0 | Female | 29.0 | False | False | False | False |
| 96451.0 | 59.0 | 82.0 | Female | 80.0 | False | False | False | False |
| 97730.0 | 62.0 | 82.0 | Female | 19.0 | False | False | False | False |
| 99828.0 | 62.0 | 85.0 | Female | 16.0 | False | False | False | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102915.0 | 75.0 | 484.0 | Female | 14.0 | True | False | False | False |
| 102926.0 | 75.0 | 484.0 | Female | 15.0 | True | False | False | False |
| 102941.0 | 75.0 | 484.0 | Female | 14.0 | True | False | False | False |
| 102945.0 | 75.0 | 484.0 | Male | 15.0 | True | False | False | False |
| 102955.0 | 75.0 | 484.0 | Female | 14.0 | True | False | False | False |
7157 rows × 8 columns
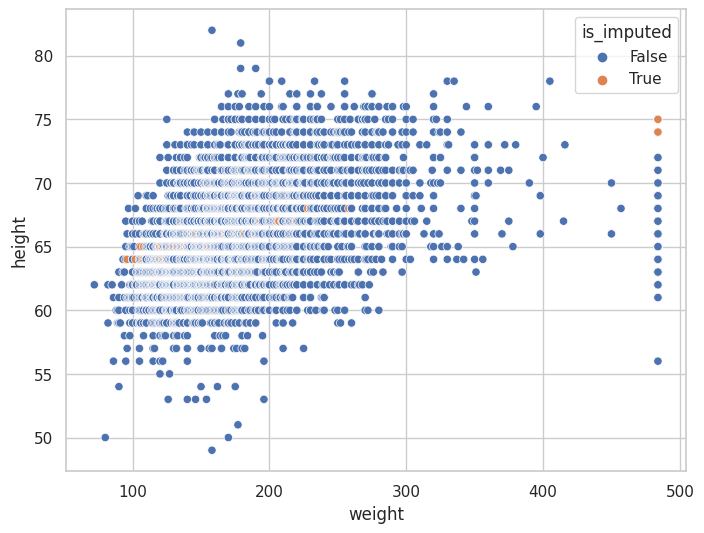
(
nhanes_model_df
.missing.scatter_imputation_plot(
x = "weight",
y = "height"
)
)
<AxesSubplot: xlabel='weight', ylabel='height'>
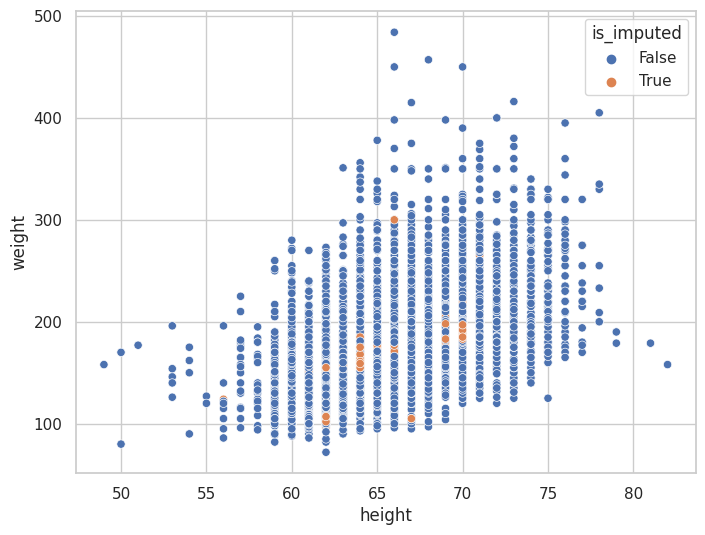
Imputaciones Múltiples por Ecuaciones Encadenadas (MICE)#
mice_imputer = sklearn.impute.IterativeImputer(
estimator=BayesianRidge(),
initial_strategy="mean",
imputation_order="ascending"
)
nhanes_mice_df = nhanes_transformed_df.copy(deep=True)
nhanes_mice_df.iloc[:, :] = mice_imputer.fit_transform(nhanes_transformed_df).round()
nhanes_mice_df = pd.concat(
[
nhanes_mice_df,
nhanes_df.missing.create_shadow_matrix(True, False, suffix="_imp")
],
axis=1
)
nhanes_mice_df
| general_health_condition | gender | height | weight | total_cholesterol | pulse | diabetes | age | height_imp | weight_imp | general_health_condition_imp | total_cholesterol_imp | pulse_imp | diabetes_imp | age_imp | gender_imp | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||||||||||
| 93705.0 | 2.0 | 0.0 | 63.0 | 165.0 | 157.0 | 52.0 | 0.0 | 66.0 | False | False | False | False | False | False | False | False |
| 93706.0 | 4.0 | 1.0 | 68.0 | 145.0 | 148.0 | 82.0 | 0.0 | 18.0 | False | False | False | False | False | False | False | False |
| 93707.0 | 2.0 | 1.0 | 70.0 | 200.0 | 189.0 | 100.0 | 0.0 | 13.0 | True | True | False | False | False | False | False | False |
| 93709.0 | 2.0 | 0.0 | 62.0 | 200.0 | 176.0 | 74.0 | 0.0 | 75.0 | False | False | True | False | False | False | False | False |
| 93711.0 | 4.0 | 1.0 | 69.0 | 142.0 | 238.0 | 62.0 | 0.0 | 56.0 | False | False | False | False | False | False | False | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | 0.0 | 1.0 | 72.0 | 180.0 | 201.0 | 96.0 | 0.0 | 33.0 | False | False | False | False | False | False | False | False |
| 102953.0 | 1.0 | 1.0 | 65.0 | 218.0 | 182.0 | 78.0 | 0.0 | 42.0 | False | False | False | False | False | False | False | False |
| 102954.0 | 2.0 | 0.0 | 66.0 | 150.0 | 172.0 | 78.0 | 0.0 | 41.0 | False | False | False | False | False | False | False | False |
| 102955.0 | 4.0 | 0.0 | 64.0 | 159.0 | 150.0 | 74.0 | 0.0 | 14.0 | True | True | False | False | False | False | False | False |
| 102956.0 | 2.0 | 1.0 | 69.0 | 250.0 | 163.0 | 76.0 | 0.0 | 38.0 | False | False | False | False | False | False | False | False |
7157 rows × 16 columns
nhanes_mice_df.missing.scatter_imputation_plot(
x = "height",
y = "weight"
)
<AxesSubplot: xlabel='height', ylabel='weight'>
Transformación inversa de los datos#
Deshacer la transformacion en dummy
nhanes_imputed_df = nhanes_mice_df.copy(deep=True)
nhanes_imputed_df[categorical_columns] = (
categorical_transformer
.named_transformers_
.ordinalencoder
.inverse_transform(
X = nhanes_mice_df[categorical_columns]
)
)
nhanes_imputed_df
| general_health_condition | gender | height | weight | total_cholesterol | pulse | diabetes | age | height_imp | weight_imp | general_health_condition_imp | total_cholesterol_imp | pulse_imp | diabetes_imp | age_imp | gender_imp | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SEQN | ||||||||||||||||
| 93705.0 | Good | Female | 63.0 | 165.0 | 157.0 | 52.0 | 0.0 | 66.0 | False | False | False | False | False | False | False | False |
| 93706.0 | Very good | Male | 68.0 | 145.0 | 148.0 | 82.0 | 0.0 | 18.0 | False | False | False | False | False | False | False | False |
| 93707.0 | Good | Male | 70.0 | 200.0 | 189.0 | 100.0 | 0.0 | 13.0 | True | True | False | False | False | False | False | False |
| 93709.0 | Good | Female | 62.0 | 200.0 | 176.0 | 74.0 | 0.0 | 75.0 | False | False | True | False | False | False | False | False |
| 93711.0 | Very good | Male | 69.0 | 142.0 | 238.0 | 62.0 | 0.0 | 56.0 | False | False | False | False | False | False | False | False |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 102949.0 | Excellent | Male | 72.0 | 180.0 | 201.0 | 96.0 | 0.0 | 33.0 | False | False | False | False | False | False | False | False |
| 102953.0 | Fair or | Male | 65.0 | 218.0 | 182.0 | 78.0 | 0.0 | 42.0 | False | False | False | False | False | False | False | False |
| 102954.0 | Good | Female | 66.0 | 150.0 | 172.0 | 78.0 | 0.0 | 41.0 | False | False | False | False | False | False | False | False |
| 102955.0 | Very good | Female | 64.0 | 159.0 | 150.0 | 74.0 | 0.0 | 14.0 | True | True | False | False | False | False | False | False |
| 102956.0 | Good | Male | 69.0 | 250.0 | 163.0 | 76.0 | 0.0 | 38.0 | False | False | False | False | False | False | False | False |
7157 rows × 16 columns
Comparativa de valores antes de imputacion con valores despues de imputacion
nhanes_df.general_health_condition.value_counts()
Good 2383
Very good 1503
Fair or 1130
Excellent 612
Poor? 169
Name: general_health_condition, dtype: int64
nhanes_imputed_df.general_health_condition.value_counts()
Good 3743
Very good 1503
Fair or 1130
Excellent 612
Poor? 169
Name: general_health_condition, dtype: int64
Me quedan entonces valores faltantes?
nhanes_imputed_df.missing.number_missing()
0
