Manejo de Datos Faltantes: Detección#
Preparacion de entorno#
pip install -r requirements.txt
^C
Requirement already satisfied: pip in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 1)) (23.1.2)
Collecting pyjanitor (from -r requirements.txt (line 2))
Using cached pyjanitor-0.25.0-py3-none-any.whl (171 kB)
Collecting missingno (from -r requirements.txt (line 3))
Using cached missingno-0.5.2-py3-none-any.whl (8.7 kB)
Requirement already satisfied: numpy in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 4)) (1.25.2)
Collecting matplotlib==3.5.1 (from -r requirements.txt (line 5))
Using cached matplotlib-3.5.1.tar.gz (35.3 MB)
Preparing metadata (setup.py): started
Preparing metadata (setup.py): finished with status 'done'
Requirement already satisfied: pandas in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 6)) (2.1.0)
Collecting pyreadr (from -r requirements.txt (line 7))
Using cached pyreadr-0.4.9-cp311-cp311-win_amd64.whl (1.3 MB)
Requirement already satisfied: seaborn in c:\users\admin\miniconda3\lib\site-packages (from -r requirements.txt (line 8)) (0.12.2)
Collecting session-info (from -r requirements.txt (line 9))
Using cached session_info-1.0.0.tar.gz (24 kB)
Preparing metadata (setup.py): started
Preparing metadata (setup.py): finished with status 'done'
Collecting upsetplot==0.6.1 (from -r requirements.txt (line 10))
Using cached UpSetPlot-0.6.1.tar.gz (18 kB)
Preparing metadata (setup.py): started
Preparing metadata (setup.py): finished with status 'done'
Requirement already satisfied: cycler>=0.10 in c:\users\admin\miniconda3\lib\site-packages (from matplotlib==3.5.1->-r requirements.txt (line 5)) (0.11.0)
Requirement already satisfied: fonttools>=4.22.0 in c:\users\admin\miniconda3\lib\site-packages (from matplotlib==3.5.1->-r requirements.txt (line 5)) (4.42.1)
Requirement already satisfied: kiwisolver>=1.0.1 in c:\users\admin\miniconda3\lib\site-packages (from matplotlib==3.5.1->-r requirements.txt (line 5)) (1.4.5)
Requirement already satisfied: packaging>=20.0 in c:\users\admin\miniconda3\lib\site-packages (from matplotlib==3.5.1->-r requirements.txt (line 5)) (23.0)
Requirement already satisfied: pillow>=6.2.0 in c:\users\admin\miniconda3\lib\site-packages (from matplotlib==3.5.1->-r requirements.txt (line 5)) (10.0.0)
Requirement already satisfied: pyparsing>=2.2.1 in c:\users\admin\miniconda3\lib\site-packages (from matplotlib==3.5.1->-r requirements.txt (line 5)) (3.0.9)
Requirement already satisfied: python-dateutil>=2.7 in c:\users\admin\miniconda3\lib\site-packages (from matplotlib==3.5.1->-r requirements.txt (line 5)) (2.8.2)
Collecting natsort (from pyjanitor->-r requirements.txt (line 2))
Downloading natsort-8.4.0-py3-none-any.whl (38 kB)
Requirement already satisfied: pandas-flavor in c:\users\admin\miniconda3\lib\site-packages (from pyjanitor->-r requirements.txt (line 2)) (0.6.0)
Collecting multipledispatch (from pyjanitor->-r requirements.txt (line 2))
Downloading multipledispatch-1.0.0-py3-none-any.whl (12 kB)
Requirement already satisfied: scipy in c:\users\admin\miniconda3\lib\site-packages (from pyjanitor->-r requirements.txt (line 2)) (1.11.2)
Requirement already satisfied: pytz>=2020.1 in c:\users\admin\miniconda3\lib\site-packages (from pandas->-r requirements.txt (line 6)) (2023.3.post1)
Requirement already satisfied: tzdata>=2022.1 in c:\users\admin\miniconda3\lib\site-packages (from pandas->-r requirements.txt (line 6)) (2023.3)
Collecting stdlib_list (from session-info->-r requirements.txt (line 9))
Downloading stdlib_list-0.9.0-py3-none-any.whl (75 kB)
0.0/75.6 kB ? eta -:--:--
---------------------------------------- 75.6/75.6 kB 2.1 MB/s eta 0:00:00
Requirement already satisfied: six>=1.5 in c:\users\admin\miniconda3\lib\site-packages (from python-dateutil>=2.7->matplotlib==3.5.1->-r requirements.txt (line 5)) (1.16.0)
Requirement already satisfied: xarray in c:\users\admin\miniconda3\lib\site-packages (from pandas-flavor->pyjanitor->-r requirements.txt (line 2)) (2023.8.0)
Building wheels for collected packages: matplotlib, upsetplot, session-info
Building wheel for matplotlib (setup.py): started
Note: you may need to restart the kernel to use updated packages.
Importar librerías
import janitor
import matplotlib.pyplot as plt
import missingno
import numpy as np
import pandas as pd
import pyreadr
import seaborn as sns
import session_info
import upsetplot
Importar funciones personalizadas#
%run pandas-missing-extension.ipynb
Configurar el aspecto general de las gráficas del proyecto
%matplotlib inline
sns.set(
rc={
"figure.figsize": (10, 10)
}
)
sns.set_style("whitegrid")
Cargar los conjuntos de datos#
Pima Indians Diabetes#
# Saving the link into a variable
pima_indians_diabetes_url = "https://nrvis.com/data/mldata/pima-indians-diabetes.csv"
# Downloading the csv file
!wget -O ./data/pima-indians-diabetes.csv { pima_indians_diabetes_url } -q
diabetes_df = pd.read_csv(
"./data/pima-indians-diabetes.csv",
sep=",",
names=[
"pregnancies",
"glucose",
"blood_pressure",
"skin_thickness",
"insulin",
"bmi",
"diabetes_pedigree_function",
"age",
"outcome",
]
)
diabetes_df.head()
| pregnancies | glucose | blood_pressure | skin_thickness | insulin | bmi | diabetes_pedigree_function | age | outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
naniar (oceanbuoys, pedestrian, riskfactors)#
Crear unidades de información de los conjuntos de datos#
base_url = "https://github.com/njtierney/naniar/raw/master/data/"
dataset_names = ("oceanbuoys", "pedestrian", "riskfactors")
extension = ".rda"
Descargar y cargar los conjuntos de datos#
datasets_dfs = {}
for dataset_name in dataset_names:
dataset_file = f"{ dataset_name }{ extension }"
dataset_output_file = f"./data/{ dataset_file }"
dataset_url = f"{ base_url }{ dataset_file }"
!wget -q -O { dataset_output_file } { dataset_url }
datasets_dfs[f"{ dataset_name }_df"] = pyreadr.read_r(dataset_output_file).get(dataset_name)
datasets_dfs.keys()
dict_keys(['oceanbuoys_df', 'pedestrian_df', 'riskfactors_df'])
Incluir conjuntos de datos en nuestro ambiente local#
locals().update(**datasets_dfs)
del datasets_dfs
Verificar carga#
oceanbuoys_df.shape, pedestrian_df.shape, riskfactors_df.shape, diabetes_df.shape
((736, 8), (37700, 9), (245, 34), (768, 9))
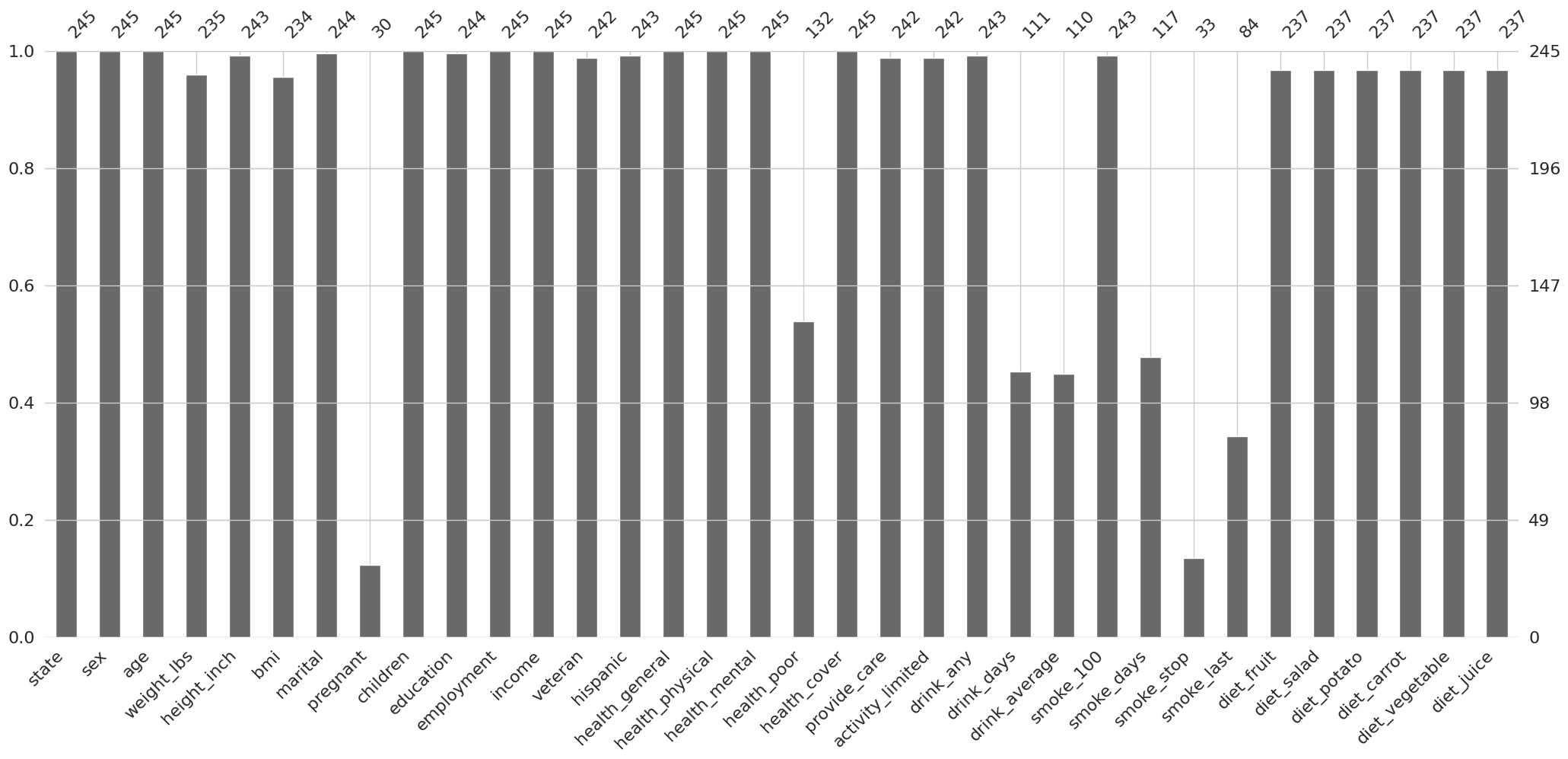
Note que la variable state tiene 245 valores no nulos, mientras que la variable pregnant tiene 30
la variable pregnant parece tener muchos valores no nulos, vamos a investigar?
riskfactors_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 245 entries, 0 to 244
Data columns (total 34 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 state 245 non-null category
1 sex 245 non-null category
2 age 245 non-null int32
3 weight_lbs 235 non-null object
4 height_inch 243 non-null object
5 bmi 234 non-null float64
6 marital 244 non-null category
7 pregnant 30 non-null category
8 children 245 non-null int32
9 education 244 non-null category
10 employment 245 non-null category
11 income 245 non-null category
12 veteran 242 non-null category
13 hispanic 243 non-null category
14 health_general 245 non-null category
15 health_physical 245 non-null int32
16 health_mental 245 non-null int32
17 health_poor 132 non-null object
18 health_cover 245 non-null category
19 provide_care 242 non-null category
20 activity_limited 242 non-null category
21 drink_any 243 non-null category
22 drink_days 111 non-null object
23 drink_average 110 non-null object
24 smoke_100 243 non-null category
25 smoke_days 117 non-null category
26 smoke_stop 33 non-null category
27 smoke_last 84 non-null category
28 diet_fruit 237 non-null object
29 diet_salad 237 non-null object
30 diet_potato 237 non-null object
31 diet_carrot 237 non-null object
32 diet_vegetable 237 non-null object
33 diet_juice 237 non-null object
dtypes: category(18), float64(1), int32(4), object(11)
memory usage: 36.7+ KB
Tabulación de valores faltantes#
Número total de valores completos (sin observaciones faltantes)#
riskfactors_df.missing.number_complete()
7144
Número total de valores faltantes#
riskfactors_df.missing.number_missing()
1186
Resúmenes tabulares de valores faltantes#
Variables / Columnas#
Resumen por variable#
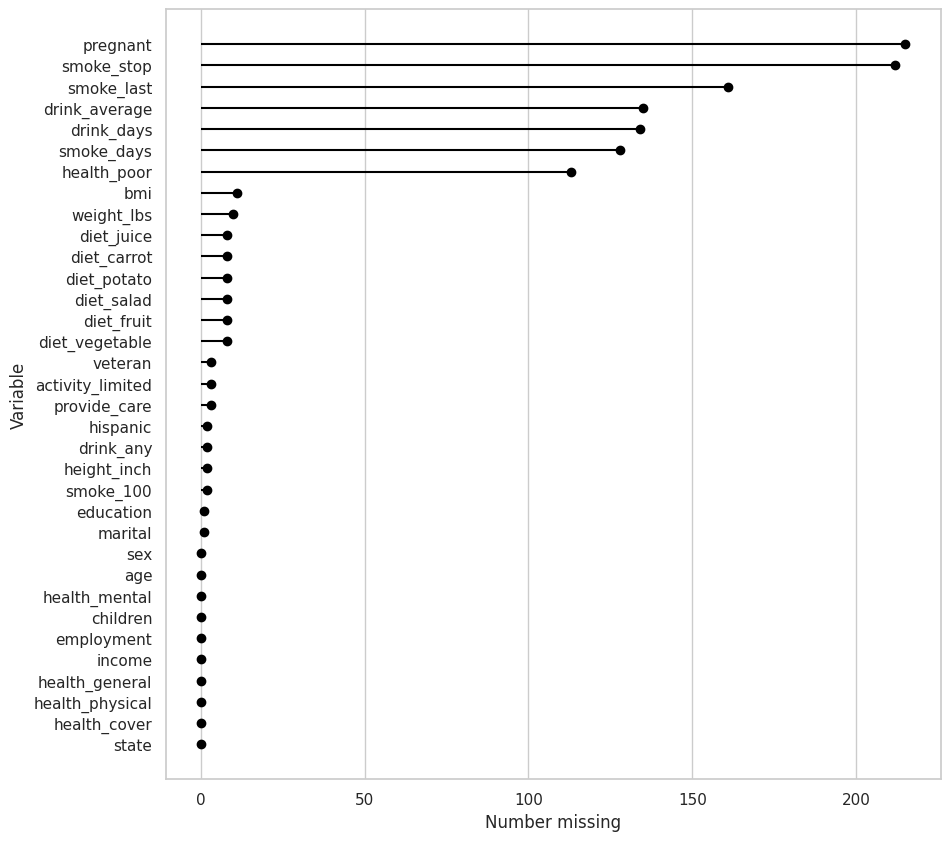
riskfactors_df.missing.missing_variable_summary()
| variable | n_missing | n_cases | pct_missing | |
|---|---|---|---|---|
| 0 | state | 0 | 245 | 0.000000 |
| 1 | sex | 0 | 245 | 0.000000 |
| 2 | age | 0 | 245 | 0.000000 |
| 3 | weight_lbs | 10 | 245 | 4.081633 |
| 4 | height_inch | 2 | 245 | 0.816327 |
| 5 | bmi | 11 | 245 | 4.489796 |
| 6 | marital | 1 | 245 | 0.408163 |
| 7 | pregnant | 215 | 245 | 87.755102 |
| 8 | children | 0 | 245 | 0.000000 |
| 9 | education | 1 | 245 | 0.408163 |
| 10 | employment | 0 | 245 | 0.000000 |
| 11 | income | 0 | 245 | 0.000000 |
| 12 | veteran | 3 | 245 | 1.224490 |
| 13 | hispanic | 2 | 245 | 0.816327 |
| 14 | health_general | 0 | 245 | 0.000000 |
| 15 | health_physical | 0 | 245 | 0.000000 |
| 16 | health_mental | 0 | 245 | 0.000000 |
| 17 | health_poor | 113 | 245 | 46.122449 |
| 18 | health_cover | 0 | 245 | 0.000000 |
| 19 | provide_care | 3 | 245 | 1.224490 |
| 20 | activity_limited | 3 | 245 | 1.224490 |
| 21 | drink_any | 2 | 245 | 0.816327 |
| 22 | drink_days | 134 | 245 | 54.693878 |
| 23 | drink_average | 135 | 245 | 55.102041 |
| 24 | smoke_100 | 2 | 245 | 0.816327 |
| 25 | smoke_days | 128 | 245 | 52.244898 |
| 26 | smoke_stop | 212 | 245 | 86.530612 |
| 27 | smoke_last | 161 | 245 | 65.714286 |
| 28 | diet_fruit | 8 | 245 | 3.265306 |
| 29 | diet_salad | 8 | 245 | 3.265306 |
| 30 | diet_potato | 8 | 245 | 3.265306 |
| 31 | diet_carrot | 8 | 245 | 3.265306 |
| 32 | diet_vegetable | 8 | 245 | 3.265306 |
| 33 | diet_juice | 8 | 245 | 3.265306 |
Tabulación del resumen por variable#
riskfactors_df.missing.missing_variable_table()
| n_missing_in_variable | n_variables | pct_variables | |
|---|---|---|---|
| 0 | 0 | 10 | 29.411765 |
| 1 | 8 | 6 | 17.647059 |
| 2 | 2 | 4 | 11.764706 |
| 3 | 3 | 3 | 8.823529 |
| 4 | 1 | 2 | 5.882353 |
| 5 | 10 | 1 | 2.941176 |
| 6 | 11 | 1 | 2.941176 |
| 7 | 113 | 1 | 2.941176 |
| 8 | 128 | 1 | 2.941176 |
| 9 | 134 | 1 | 2.941176 |
| 10 | 135 | 1 | 2.941176 |
| 11 | 161 | 1 | 2.941176 |
| 12 | 212 | 1 | 2.941176 |
| 13 | 215 | 1 | 2.941176 |
Casos / Observaciones / Filas#
Resúmenes por caso (muestra valores nulos por row)#
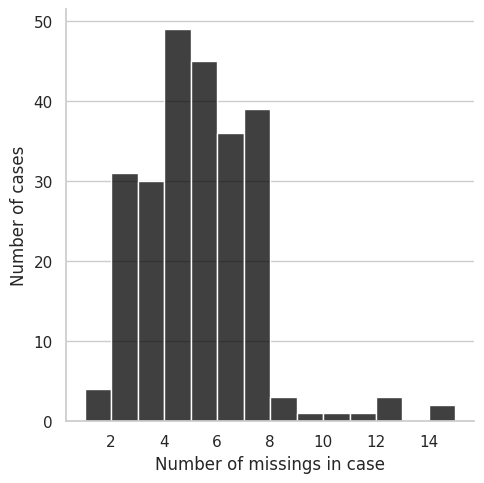
riskfactors_df.missing.missing_case_summary()
| case | n_missing | pct_missing | |
|---|---|---|---|
| 0 | 0 | 6 | 16.666667 |
| 1 | 1 | 6 | 16.666667 |
| 2 | 2 | 7 | 19.444444 |
| 3 | 3 | 12 | 33.333333 |
| 4 | 4 | 5 | 13.888889 |
| ... | ... | ... | ... |
| 240 | 240 | 6 | 16.666667 |
| 241 | 241 | 5 | 13.888889 |
| 242 | 242 | 3 | 8.333333 |
| 243 | 243 | 2 | 5.555556 |
| 244 | 244 | 3 | 8.333333 |
245 rows × 3 columns
Tabulación del resumen por caso#
riskfactors_df.missing.missing_case_table()
| n_missing_in_case | n_cases | pct_case | |
|---|---|---|---|
| 0 | 4 | 49 | 20.000000 |
| 1 | 5 | 45 | 18.367347 |
| 2 | 7 | 39 | 15.918367 |
| 3 | 6 | 36 | 14.693878 |
| 4 | 2 | 31 | 12.653061 |
| 5 | 3 | 30 | 12.244898 |
| 6 | 1 | 4 | 1.632653 |
| 7 | 8 | 3 | 1.224490 |
| 8 | 12 | 3 | 1.224490 |
| 9 | 15 | 2 | 0.816327 |
| 10 | 9 | 1 | 0.408163 |
| 11 | 10 | 1 | 0.408163 |
| 12 | 11 | 1 | 0.408163 |
Intervalos de valores faltantes#
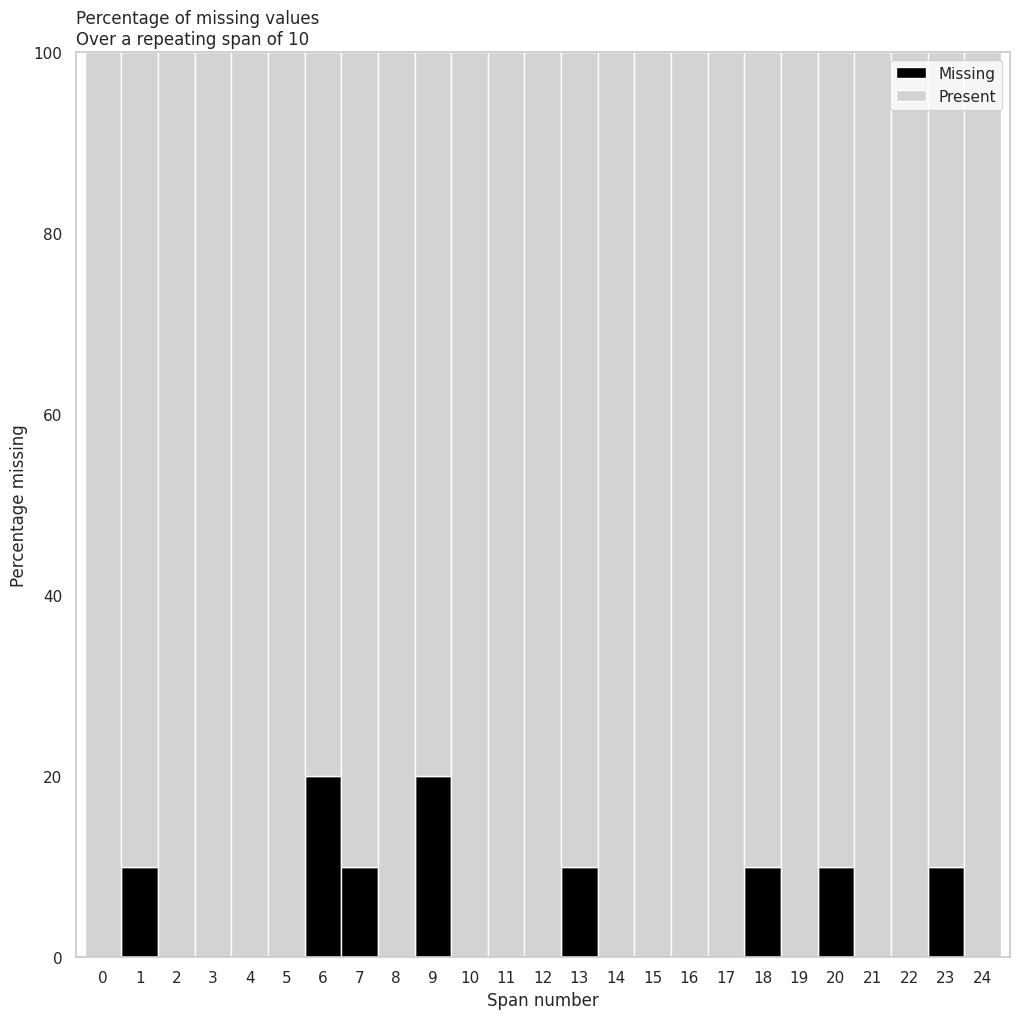
(
riskfactors_df
.missing
.missing_variable_span(
variable="weight_lbs", # variable a analizar
span_every=50 # intervalos a romper la variable
)
)
| span_counter | n_missing | n_complete | pct_missing | pct_complete | |
|---|---|---|---|---|---|
| 0 | 0 | 1 | 49 | 2.000000 | 98.000000 |
| 1 | 1 | 5 | 45 | 10.000000 | 90.000000 |
| 2 | 2 | 1 | 49 | 2.000000 | 98.000000 |
| 3 | 3 | 1 | 49 | 2.000000 | 98.000000 |
| 4 | 4 | 2 | 43 | 4.444444 | 95.555556 |
Run length de valores faltantes#
Detecta automáticamente el conteo seguido de variables nulas
(
riskfactors_df
.missing
.missing_variable_run(
variable="weight_lbs"
)
)
| run_length | is_na | |
|---|---|---|
| 0 | 14 | complete |
| 1 | 1 | missing |
| 2 | 45 | complete |
| 3 | 1 | missing |
| 4 | 5 | complete |
| 5 | 1 | missing |
| 6 | 12 | complete |
| 7 | 1 | missing |
| 8 | 10 | complete |
| 9 | 2 | missing |
| 10 | 40 | complete |
| 11 | 1 | missing |
| 12 | 53 | complete |
| 13 | 1 | missing |
| 14 | 14 | complete |
| 15 | 1 | missing |
| 16 | 31 | complete |
| 17 | 1 | missing |
| 18 | 11 | complete |
Visualización inicial de valores faltantes#
Variable#
riskfactors_df.missing.missing_variable_plot()
Casos / Observaciones / Filas#
riskfactors_df.missing.missing_case_plot()
(
riskfactors_df
.missing
.missing_variable_span_plot(
variable="weight_lbs",
span_every=10,
rot = 0 # label rotation
)
)
Que tan completas están nuestras variables?
missingno.bar(df = riskfactors_df);
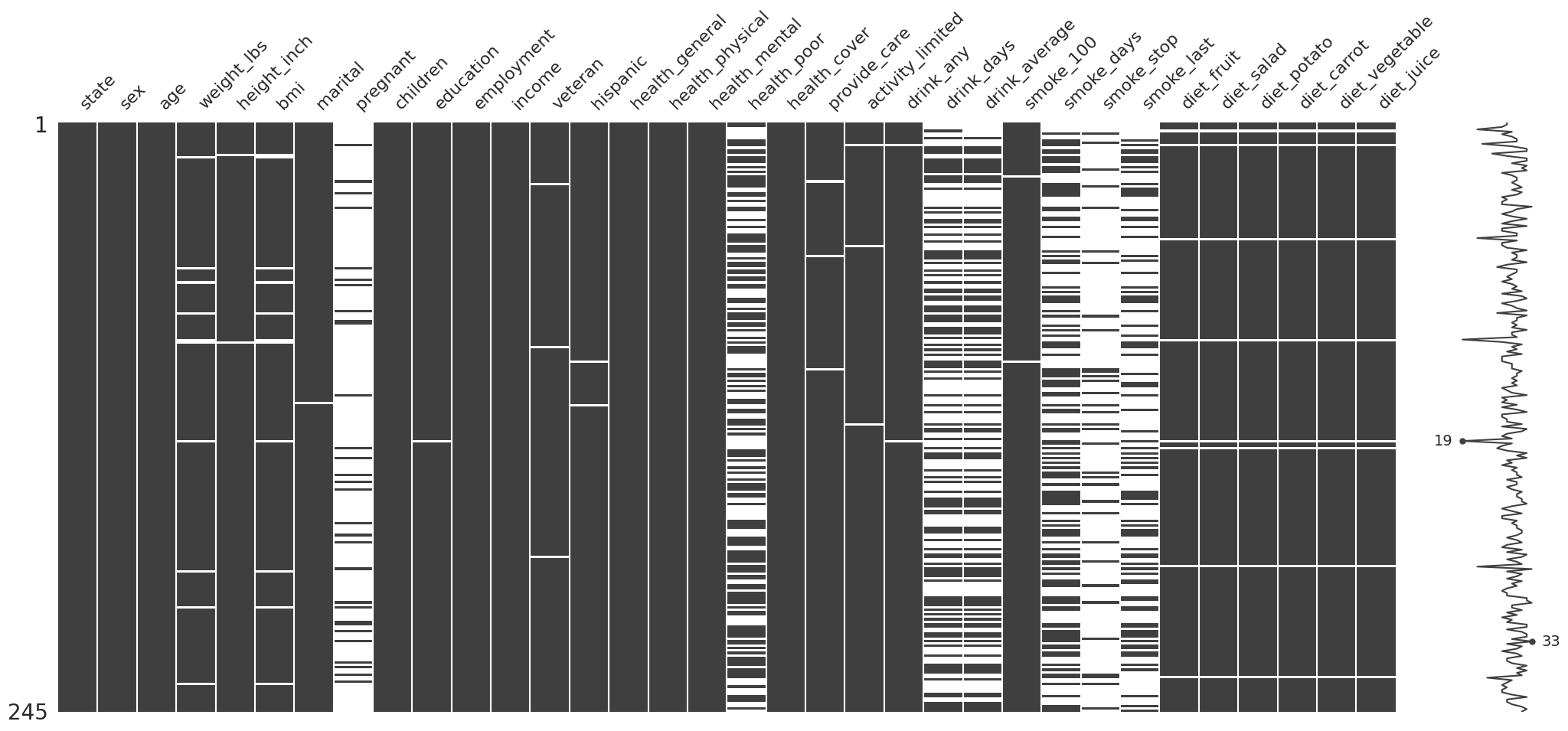
missingno.matrix(df=riskfactors_df);
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:73: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax0.grid(b=False)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:142: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax1.grid(b=False)
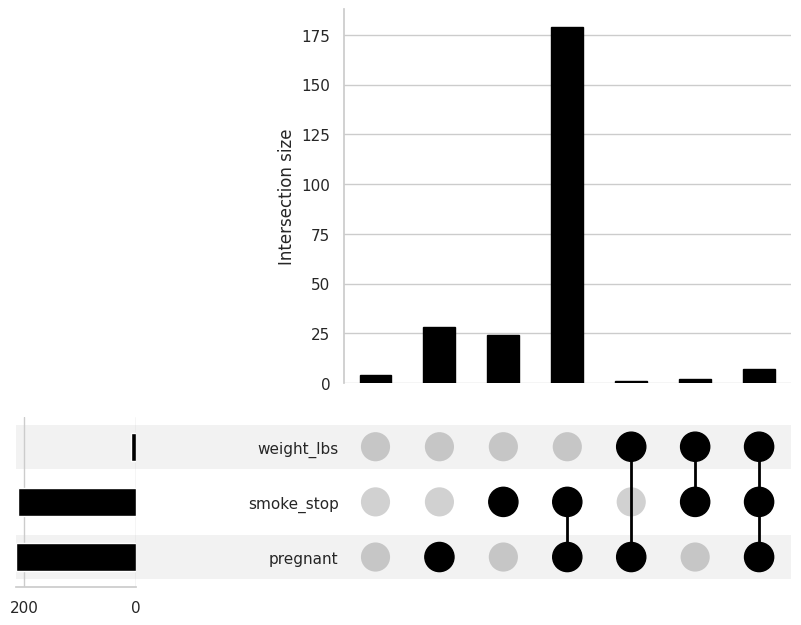
(
riskfactors_df
.missing
.missing_upsetplot(
variables=["pregnant","weight_lbs","smoke_stop"], # Put the target variable name
# alternatively, put none to see all variables
element_size = 60 # This one specifies the size of the figure that you want to create
)
);
Codificación de valores faltantes#
Al igual que cada persona es una nueva puerta a un mundo diferente, los valores faltantes existen en diferentes formas y colores. Al trabajar con valores faltantes será crítico entender sus distintas representaciones. A pesar de que el conjunto de datos de trabajo pareciera que no contiene valores faltantes, deberás ser capaz de ir más allá de lo observado a simple vista para remover el manto tras el cual se esconde lo desconocido.
Valores comúnmente asociados a valores faltantes#
Cadenas de texto#
common_na_strings = (
"missing",
"NA",
"N A",
"N/A",
"#N/A",
"NA ",
" NA",
"N /A",
"N / A",
" N / A",
"N / A ",
"na",
"n a",
"n/a",
"na ",
" na",
"n /a",
"n / a",
" a / a",
"n / a ",
"NULL",
"null",
"",
"?",
"*",
".",
)
Números#
common_na_numbers = (-9, -99, -999, -9999, 9999, 66, 77, 88, -1)
¿Cómo encontrar los valores comúnmente asociados a valores faltantes?#
missing_data_example_df = pd.DataFrame.from_dict(
dict(
x = [1, 3, "NA", -99, -98, -99],
y = ["A", "N/A", "NA", "E", "F", "G"],
z = [-100, -99, -98, -101, -1, -1]
)
)
missing_data_example_df
| x | y | z | |
|---|---|---|---|
| 0 | 1 | A | -100 |
| 1 | 3 | N/A | -99 |
| 2 | NA | NA | -98 |
| 3 | -99 | E | -101 |
| 4 | -98 | F | -1 |
| 5 | -99 | G | -1 |
Ya sabemos que hay valores faltantes
Será que Pandas los detecta?
missing_data_example_df.isna()
| x | y | z | |
|---|---|---|---|
| 0 | False | False | False |
| 1 | False | False | False |
| 2 | False | False | False |
| 3 | False | False | False |
| 4 | False | False | False |
| 5 | False | False | False |
Sustituyendo valores comúnmente asociados a valores faltantes#
Sustitución desde la lectura de datos#
pd.read_csv(
"./data/missing_data_enconding_example.csv",
na_filter=True,
na_values=[-99, -1]
)
| x | y | z | |
|---|---|---|---|
| 0 | 1.0 | A | -100.0 |
| 1 | 3.0 | NaN | NaN |
| 2 | NaN | NaN | -98.0 |
| 3 | NaN | E | -101.0 |
| 4 | -98.0 | F | NaN |
| 5 | NaN | G | NaN |
Sustitución global#
(
missing_data_example_df
.replace(
to_replace=[-99, "NA"],
value=np.nan
)
)
| x | y | z | |
|---|---|---|---|
| 0 | 1.0 | A | -100.0 |
| 1 | 3.0 | N/A | NaN |
| 2 | NaN | NaN | -98.0 |
| 3 | NaN | E | -101.0 |
| 4 | -98.0 | F | -1.0 |
| 5 | NaN | G | -1.0 |
Sustitución dirigida#
Sustituir solamente en una columna
(
missing_data_example_df
.replace(
to_replace={
"x":{-99:np.nan}
}
)
)
| x | y | z | |
|---|---|---|---|
| 0 | 1 | A | -100 |
| 1 | 3 | N/A | -99 |
| 2 | NA | NA | -98 |
| 3 | NaN | E | -101 |
| 4 | -98 | F | -1 |
| 5 | NaN | G | -1 |
Conversión de valores faltantes implícitos a explícitos#
"Implícito se refiere a todo aquello que se entiende que está incluido pero sin ser expresado de forma directa o explícitamente."
Un valor faltante implícito indica que el valor faltante debería estar incluido
en el conjunto de datos del análisis, sin que éste lo diga o lo especifique.
Por lo general, son valores que podemos encontrar al pivotar nuestros datos
o contabilizar el número de apariciones de combinaciones de las variables de estudio.
implicit_to_explicit_df = pd.DataFrame.from_dict(
data={
"name": ["lynn", "lynn", "lynn", "zelda"],
"time": ["morning", "afternoon", "night", "morning"],
"value": [350, 310, np.nan, 320]
}
)
implicit_to_explicit_df
| name | time | value | |
|---|---|---|---|
| 0 | lynn | morning | 350.0 |
| 1 | lynn | afternoon | 310.0 |
| 2 | lynn | night | NaN |
| 3 | zelda | morning | 320.0 |
Note que implicitamente hay valores faltantes, por que zelda no tiene valores para afternoon & night?
Estrategias para la identificación de valores faltantes implícitos#
Pivotar la tabla de datos#
(
implicit_to_explicit_df
.pivot_wider(
index="name",
names_from="time",
values_from="value"
)
)
| name | afternoon | morning | night | |
|---|---|---|---|---|
| 0 | lynn | 310.0 | 350.0 | NaN |
| 1 | zelda | NaN | 320.0 | NaN |
Cuantificar ocurrencias de n-tuplas#
(
implicit_to_explicit_df
.value_counts(
subset=["name"]
)
.reset_index(name="n")
.query("n<3")
)
| name | n | |
|---|---|---|
| 1 | zelda | 1 |
Exponer filas faltantes implícitas a explícitas#
janitor.complete() está modelada a partir de la función complete() del paquete tidyr y es un wrapper alrededor de janitor.expand_grid(), pd.merge() y pd.fillna(). En cierto modo, es lo contrario de pd.dropna(), ya que expone implícitamente las filas que faltan.
Son posibles combinaciones de nombres de columnas o una lista/tupla de nombres de columnas, o incluso un diccionario de nombres de columna y nuevos valores.
Las columnas MultiIndex no son complatibles.
Exponer n-tuplas de valores faltantes#
Ejemplo, encontrar los pares faltantes de name y time.
(
implicit_to_explicit_df
#janitor
.complete(
"name",
"time"
)
)
| name | time | value | |
|---|---|---|---|
| 0 | lynn | morning | 350.0 |
| 1 | lynn | afternoon | 310.0 |
| 2 | lynn | night | NaN |
| 3 | zelda | morning | 320.0 |
| 4 | zelda | afternoon | NaN |
| 5 | zelda | night | NaN |
Limitar la exposición de n-tuplas de valores faltantes#
(
implicit_to_explicit_df
#janitor
.complete(
{"name":["lynn", "zelda"]},
{"time":["morning", "afternoon"]},
sort=True
)
)
| name | time | value | |
|---|---|---|---|
| 0 | lynn | afternoon | 310.0 |
| 1 | lynn | morning | 350.0 |
| 2 | zelda | afternoon | NaN |
| 3 | zelda | morning | 320.0 |
| 4 | lynn | night | NaN |
Rellenar los valores faltantes#
(
implicit_to_explicit_df
#janitor
.complete(
"name",
"time",
fill_value = np.nan #aqui se puede poner cualquier valor para los nulos
)
)
| name | time | value | |
|---|---|---|---|
| 0 | lynn | morning | 350.0 |
| 1 | lynn | afternoon | 310.0 |
| 2 | lynn | night | NaN |
| 3 | zelda | morning | 320.0 |
| 4 | zelda | afternoon | NaN |
| 5 | zelda | night | NaN |
Limitar el rellenado de valores faltantes implícitos#
(
implicit_to_explicit_df
#janitor
.complete(
"name",
"time",
fill_value = 0,
explicit=False
)
)
| name | time | value | |
|---|---|---|---|
| 0 | lynn | morning | 350.0 |
| 1 | lynn | afternoon | 310.0 |
| 2 | lynn | night | NaN |
| 3 | zelda | morning | 320.0 |
| 4 | zelda | afternoon | 0.0 |
| 5 | zelda | night | 0.0 |
Tipos de valores faltantes#
Usando el DF de diabetes, determine si hay valores faltantes
diabetes_df.missing.missing_variable_plot()
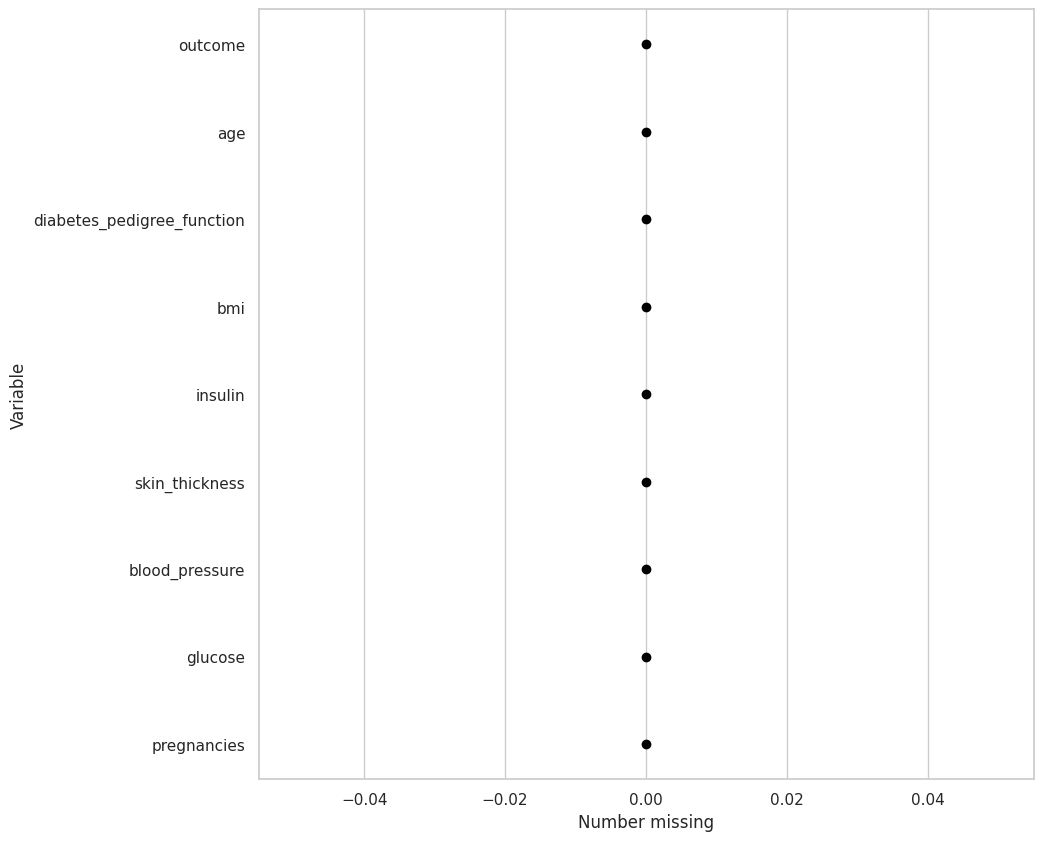
This chart is basically saying that there are no missing values, which is false. The problem is that the missing values are in a different format
The missing values are shown as a zero, let’s change them into numpy nan
diabetes_df[diabetes_df.columns[1:6]] = diabetes_df[diabetes_df.columns[1:6]].replace(0, np.nan)
diabetes_df.missing.missing_variable_plot()
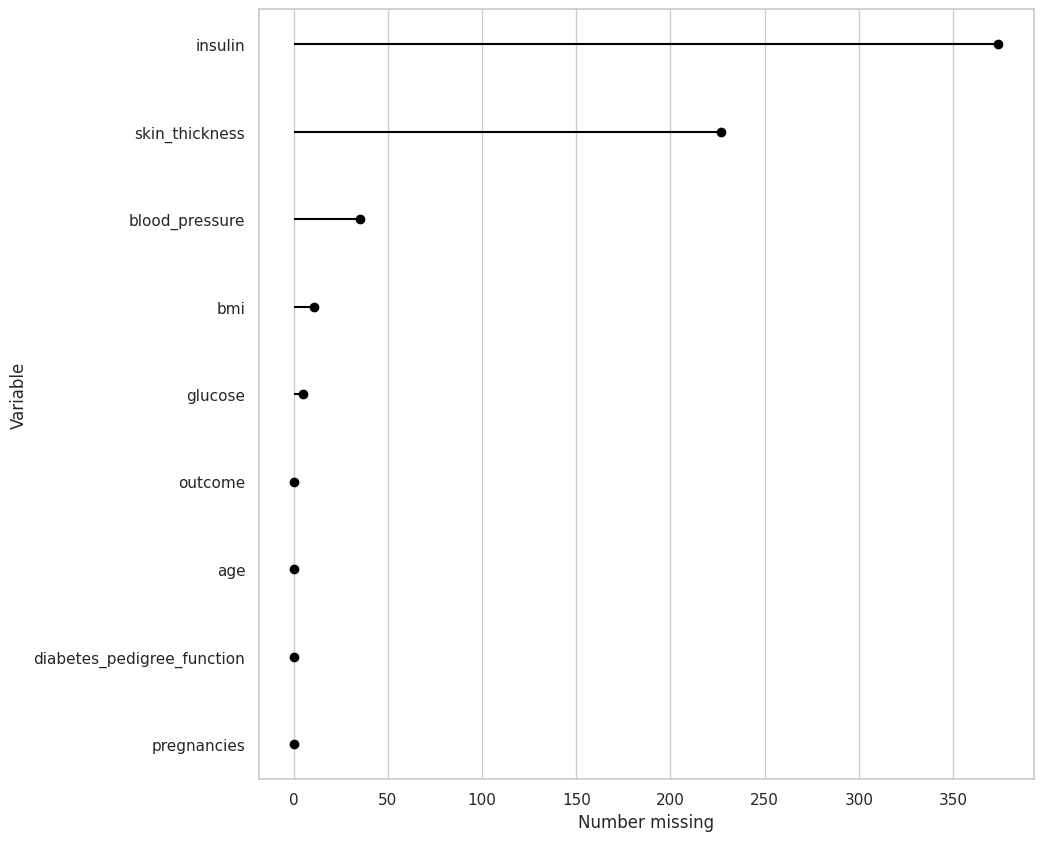
Missing Completely At Random (MCAR)#
(
diabetes_df
.missing.sort_variables_by_missingness()
.pipe(missingno.matrix)
)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:73: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax0.grid(b=False)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:142: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax1.grid(b=False)
<AxesSubplot:>
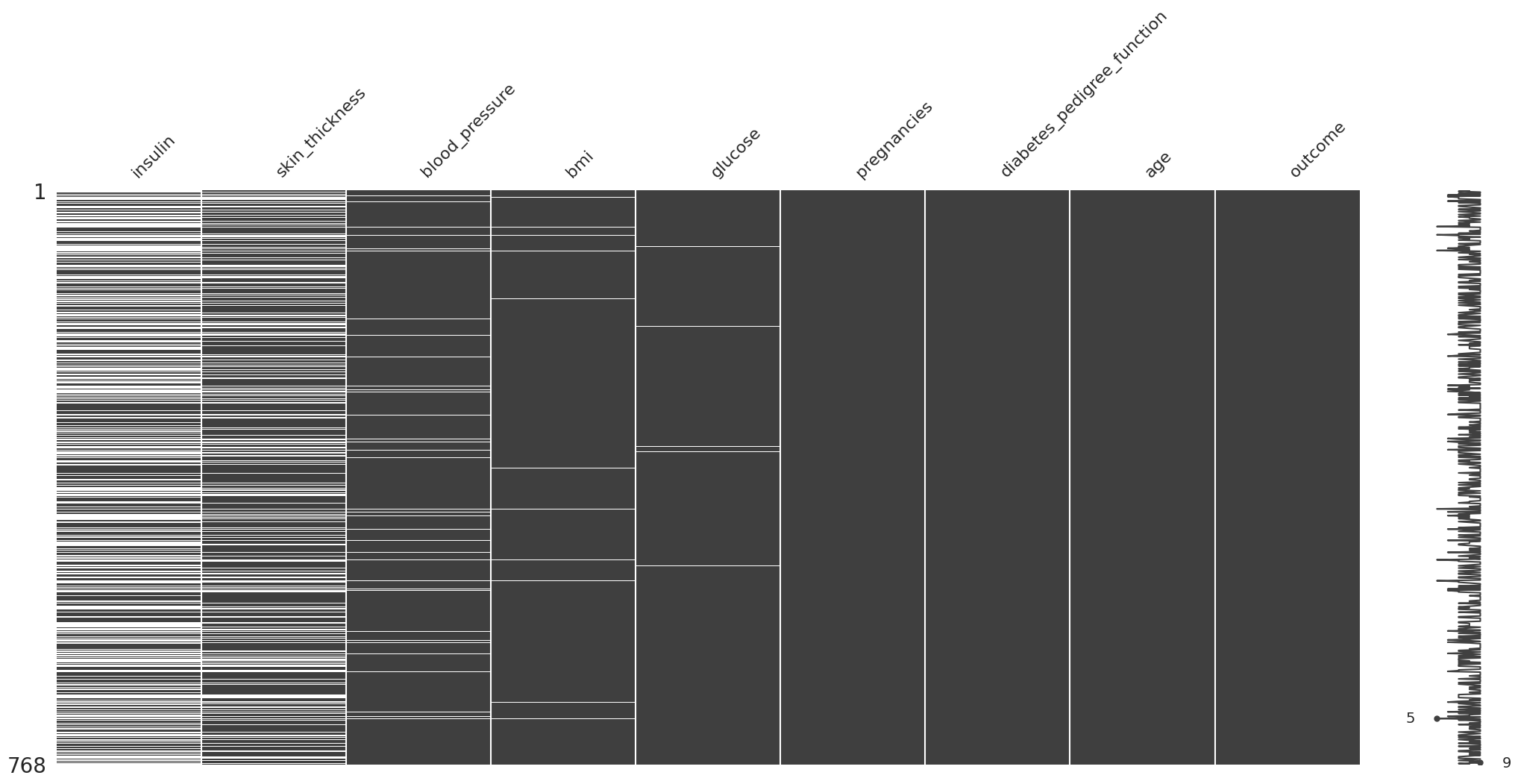
Se ordenan las columnas tal que las que tienen mas valores faltantes aparecen primero.
Missing At Random (MAR)#
(
diabetes_df
.missing.sort_variables_by_missingness()
.sort_values(by="blood_pressure")
.pipe(missingno.matrix)
)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:73: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax0.grid(b=False)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:142: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax1.grid(b=False)
<AxesSubplot:>
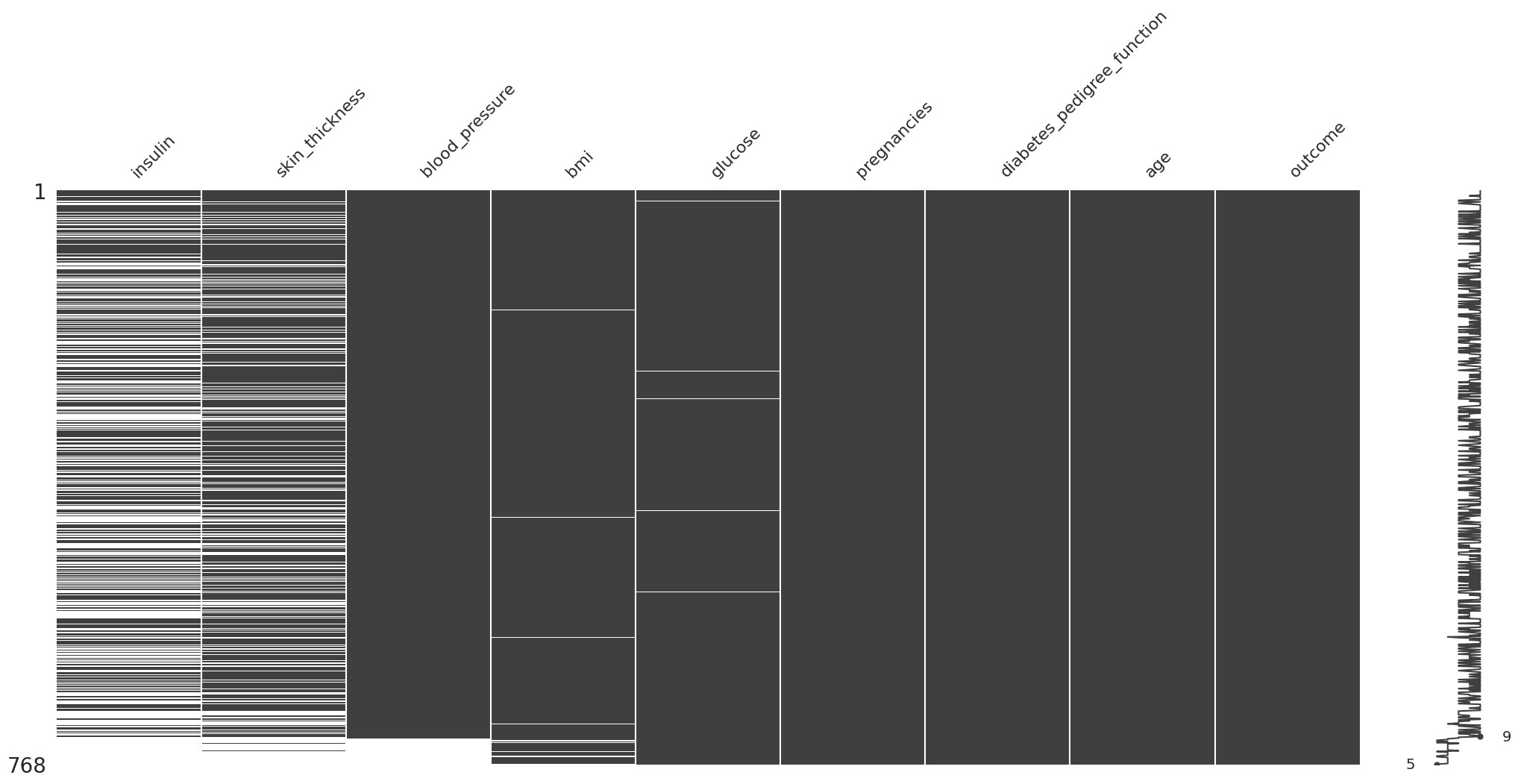
Aqui se puede empezar a inferir que puede haber una relacion entre la presion sanguinea y los datos faltantes
Missing Not At Random (MNAR)#
(
diabetes_df
.missing.sort_variables_by_missingness()
.sort_values(by="insulin")
.pipe(missingno.matrix)
)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:73: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax0.grid(b=False)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:142: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax1.grid(b=False)
<AxesSubplot:>
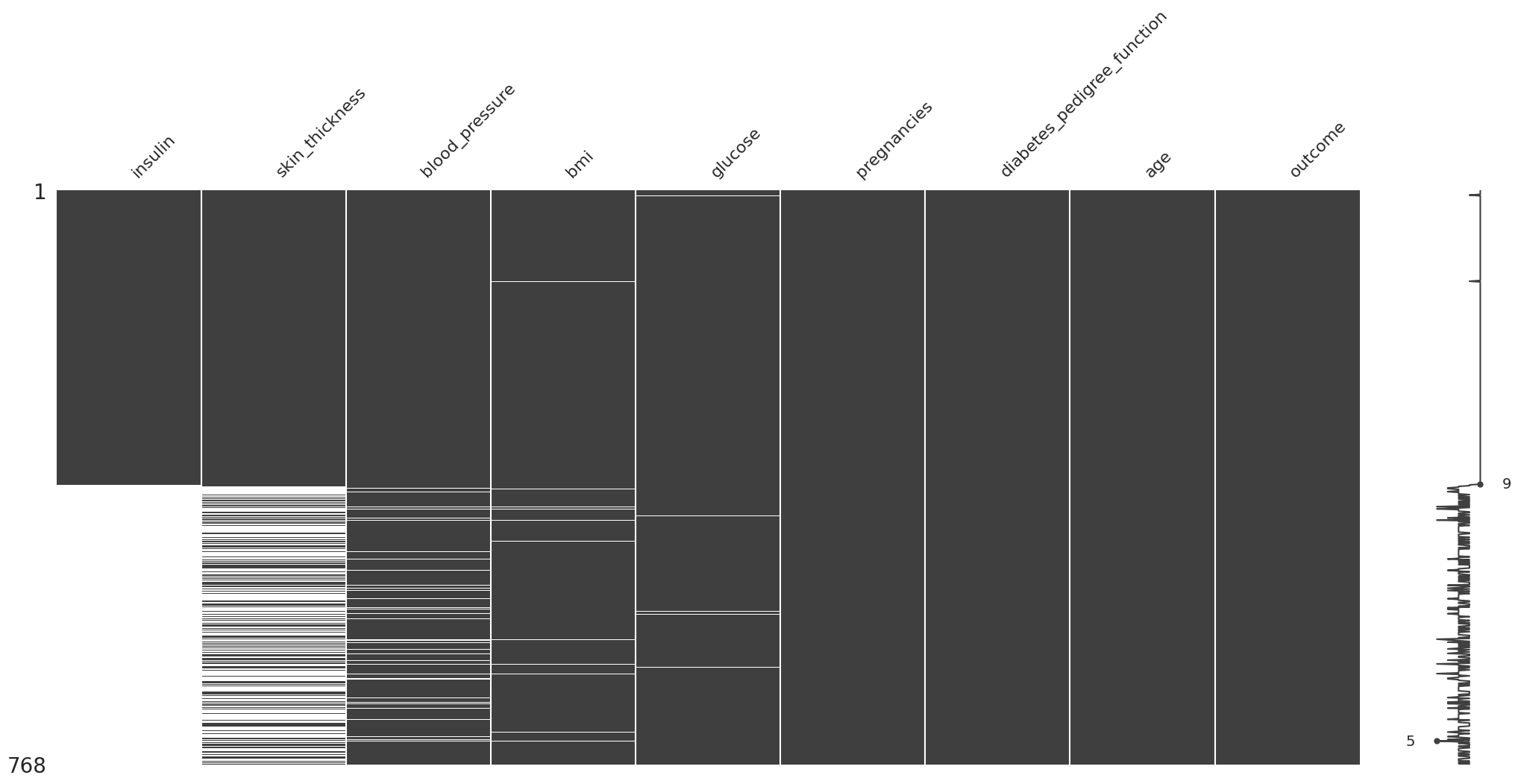
Aqui podemos observar que este error es sistematico. Siempre que se colectan los datos de insulina, tambien se colectan el resto de datos. A su vez, cuando no se colectan los datos de insulina, tienden a haber datos faltantes en las otras variables
Concepto y aplicación de la matriz de sombras (i.e., shadow matrix)#
Construcción de la matriz de sombras#
(
riskfactors_df
.isna()
.replace({
False:"Not missing",
True:"Missing"
})
.add_suffix("_NA")
.pipe(
lambda shadow_matrix: pd.concat(
[riskfactors_df, shadow_matrix],
axis="columns"
)
)
)
| state | sex | age | weight_lbs | height_inch | bmi | marital | pregnant | children | education | ... | smoke_100_NA | smoke_days_NA | smoke_stop_NA | smoke_last_NA | diet_fruit_NA | diet_salad_NA | diet_potato_NA | diet_carrot_NA | diet_vegetable_NA | diet_juice_NA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 26 | Female | 49 | 190 | 64 | 32.68 | Married | NaN | 0 | 6 | ... | Not missing | Missing | Missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
| 1 | 40 | Female | 48 | 170 | 68 | 25.90 | Divorced | NaN | 0 | 5 | ... | Not missing | Missing | Missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
| 2 | 72 | Female | 55 | 163 | 64 | 28.04 | Married | NaN | 0 | 4 | ... | Not missing | Missing | Missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
| 3 | 42 | Male | 42 | 230 | 74 | 29.59 | Married | NaN | 1 | 6 | ... | Not missing | Missing | Missing | Missing | Missing | Missing | Missing | Missing | Missing | Missing |
| 4 | 32 | Female | 66 | 135 | 62 | 24.74 | Widowed | NaN | 0 | 5 | ... | Not missing | Not missing | Not missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 240 | 10 | Female | 79 | 144 | 63 | 25.56 | Widowed | NaN | 0 | 4 | ... | Not missing | Missing | Missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
| 241 | 46 | Male | 45 | 170 | 74 | 21.87 | Divorced | NaN | 2 | 4 | ... | Not missing | Missing | Missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
| 242 | 15 | Male | 62 | 175 | 71 | 24.46 | Divorced | NaN | 0 | 6 | ... | Not missing | Not missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
| 243 | 34 | Female | 62 | 138 | 64 | 23.74 | Married | NaN | 0 | 4 | ... | Not missing | Not missing | Not missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
| 244 | 18 | Male | 9 | 200 | 70 | 28.76 | Married | NaN | 0 | 4 | ... | Not missing | Not missing | Missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing | Not missing |
245 rows × 68 columns
Utilizar función de utilería bind_shadow_matrix()#
(
riskfactors_df
.missing
.bind_shadow_matrix(only_missing=True) #Only missing=true will just concat cols with missing values
)
| state | sex | age | weight_lbs | height_inch | bmi | marital | pregnant | children | education | ... | smoke_100_NA | smoke_days_NA | smoke_stop_NA | smoke_last_NA | diet_fruit_NA | diet_salad_NA | diet_potato_NA | diet_carrot_NA | diet_vegetable_NA | diet_juice_NA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 26 | Female | 49 | 190 | 64 | 32.68 | Married | NaN | 0 | 6 | ... | Not Missing | Missing | Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
| 1 | 40 | Female | 48 | 170 | 68 | 25.90 | Divorced | NaN | 0 | 5 | ... | Not Missing | Missing | Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
| 2 | 72 | Female | 55 | 163 | 64 | 28.04 | Married | NaN | 0 | 4 | ... | Not Missing | Missing | Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
| 3 | 42 | Male | 42 | 230 | 74 | 29.59 | Married | NaN | 1 | 6 | ... | Not Missing | Missing | Missing | Missing | Missing | Missing | Missing | Missing | Missing | Missing |
| 4 | 32 | Female | 66 | 135 | 62 | 24.74 | Widowed | NaN | 0 | 5 | ... | Not Missing | Not Missing | Not Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 240 | 10 | Female | 79 | 144 | 63 | 25.56 | Widowed | NaN | 0 | 4 | ... | Not Missing | Missing | Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
| 241 | 46 | Male | 45 | 170 | 74 | 21.87 | Divorced | NaN | 2 | 4 | ... | Not Missing | Missing | Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
| 242 | 15 | Male | 62 | 175 | 71 | 24.46 | Divorced | NaN | 0 | 6 | ... | Not Missing | Not Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
| 243 | 34 | Female | 62 | 138 | 64 | 23.74 | Married | NaN | 0 | 4 | ... | Not Missing | Not Missing | Not Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
| 244 | 18 | Male | 9 | 200 | 70 | 28.76 | Married | NaN | 0 | 4 | ... | Not Missing | Not Missing | Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing | Not Missing |
245 rows × 58 columns
Explorar estadísticos utilizando las nuevas columnas de la matriz de sombras#
(
riskfactors_df
.missing
.bind_shadow_matrix(only_missing=True)
.groupby(["weight_lbs_NA"])
["age"]
.describe()
.reset_index()
)
| weight_lbs_NA | count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Missing | 10.0 | 60.100000 | 13.706851 | 37.0 | 52.25 | 62.5 | 65.0 | 82.0 |
| 1 | Not Missing | 235.0 | 58.021277 | 17.662904 | 7.0 | 47.50 | 59.0 | 70.0 | 97.0 |
Visualización de valores faltantes en una variable#
(
riskfactors_df
.missing
.bind_shadow_matrix(only_missing=True)
.pipe(
lambda df:(
sns.boxenplot(
data=df,
x="weight_lbs_NA",
y="age"
)
)
)
)
<AxesSubplot:xlabel='weight_lbs_NA', ylabel='age'>
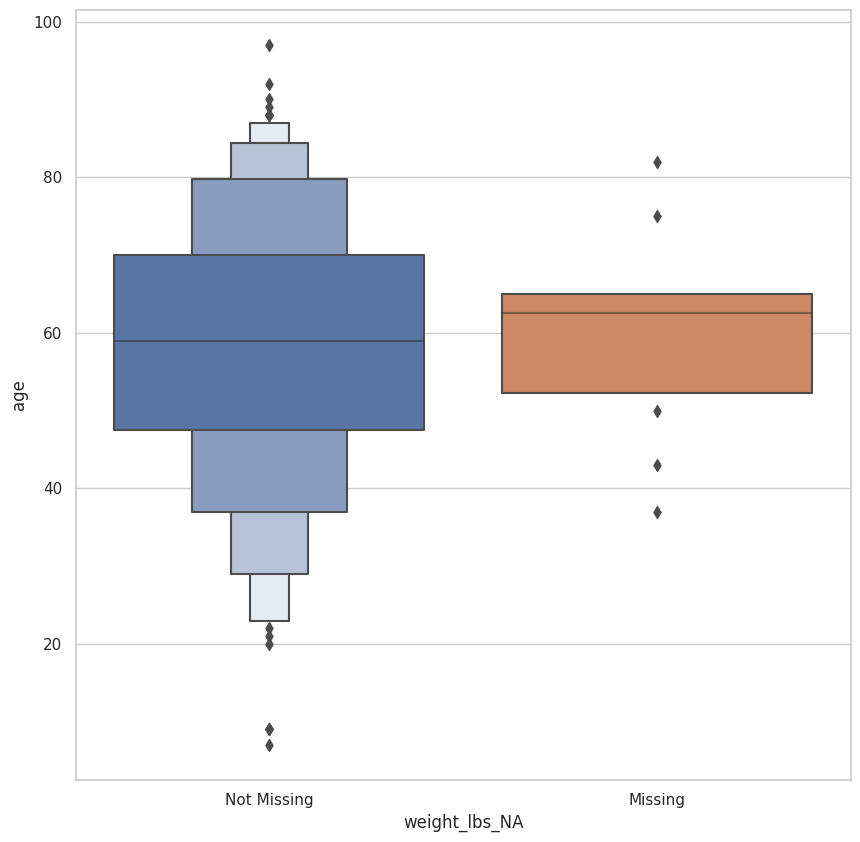
(
riskfactors_df
.missing
.bind_shadow_matrix(only_missing=True)
.pipe(
lambda df:(
sns.displot(
data=df,
x="age",
hue="weight_lbs_NA",
kind="kde"
)
)
)
)
<seaborn.axisgrid.FacetGrid at 0x7f3f7c0a0670>
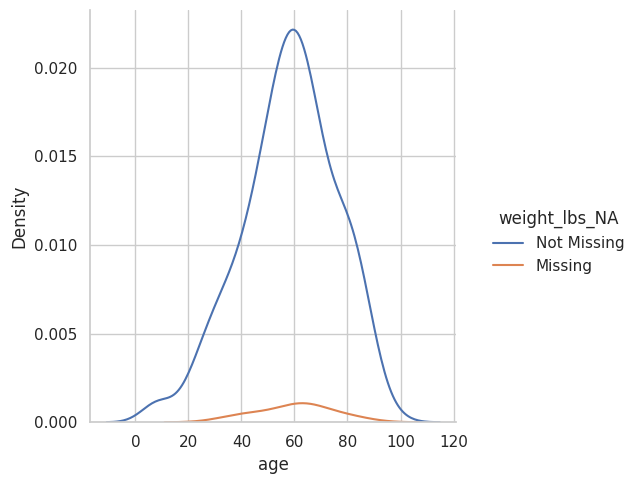
(
riskfactors_df
.missing
.bind_shadow_matrix(only_missing=True)
.pipe(
lambda df:(
sns.displot(
data=df,
x="age",
col="weight_lbs_NA",
facet_kws={
"sharey":False
}
)
)
)
)
<seaborn.axisgrid.FacetGrid at 0x7f3f7c121c70>
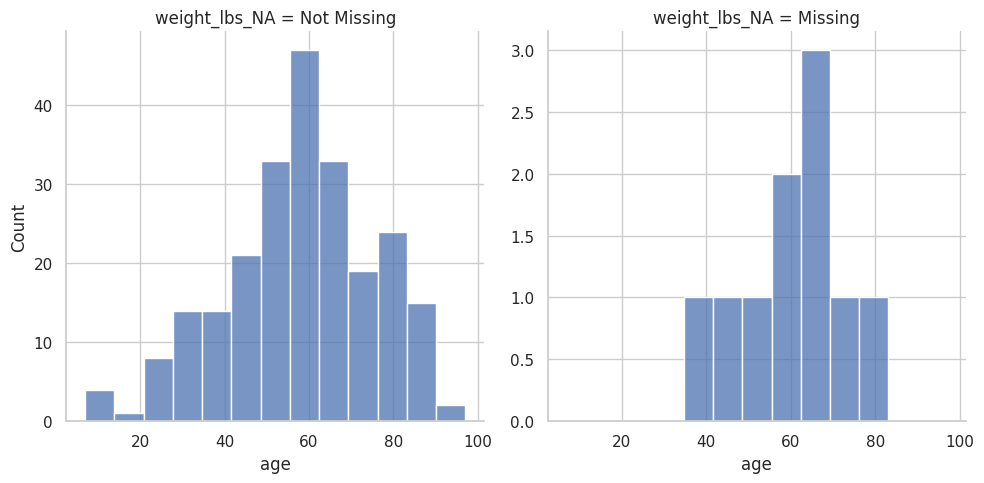
(
riskfactors_df
.missing
.bind_shadow_matrix(only_missing=True)
.pipe(
lambda df:(
sns.displot(
data=df,
x="age",
col="marital_NA",
row="weight_lbs_NA"
)
)
)
)
<seaborn.axisgrid.FacetGrid at 0x7f3f755a59a0>
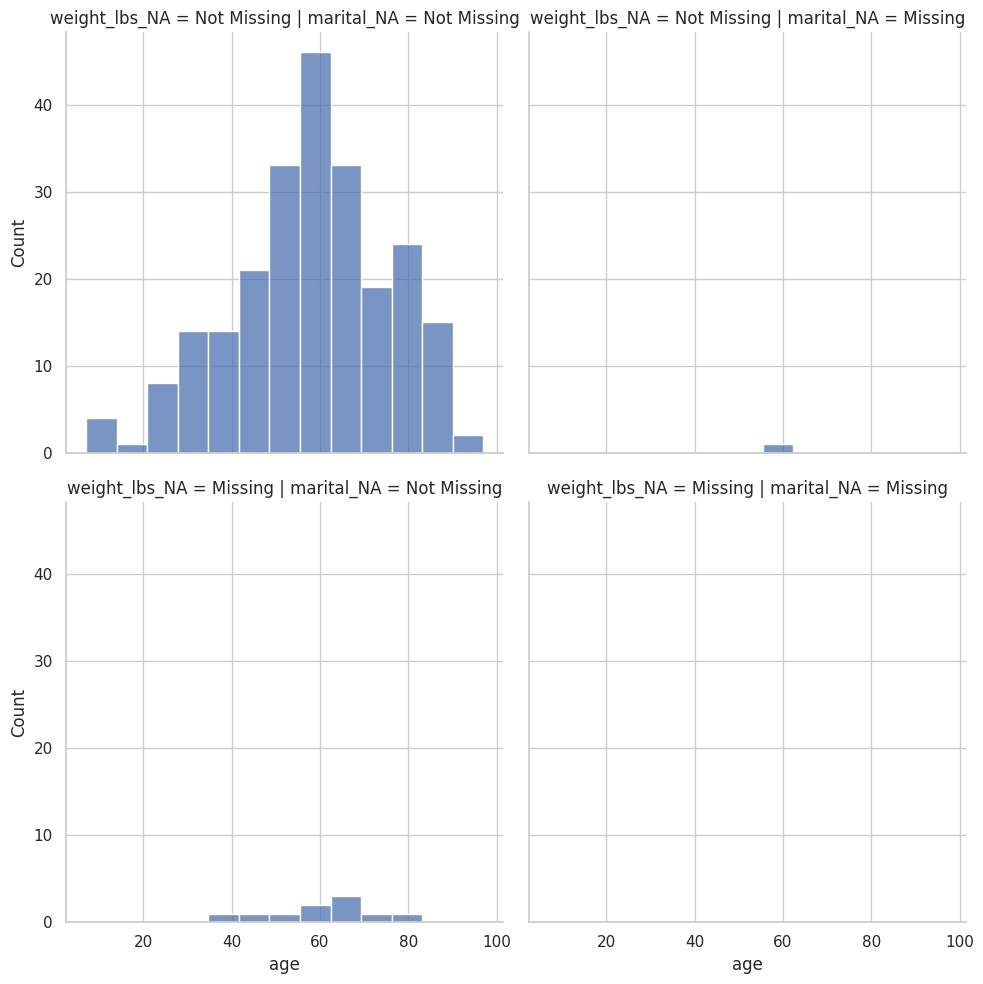
Visualización de valores faltantes en dos variables#
def column_fill_with_dummies(
column: pd.Series,
proportion_below: float=0.10,
jitter: float=0.075,
seed: int=42
) -> pd.Series:
column = column.copy(deep=True)
# Extract values metadata
missing_mask = column.isna()
number_missing_values = missing_mask.sum()
column_range = column.max() - column.min()
# Shift data
column_shift = column.min() - column.min() * proportion_below
# Create the "jitter" (noise) to be added around the points.
np.random.seed(seed)
column_jitter = (np.random.rand(number_missing_values) - 2) * column_range * jitter
# Save new dummy data.
column[missing_mask] = column_shift + column_jitter
return column
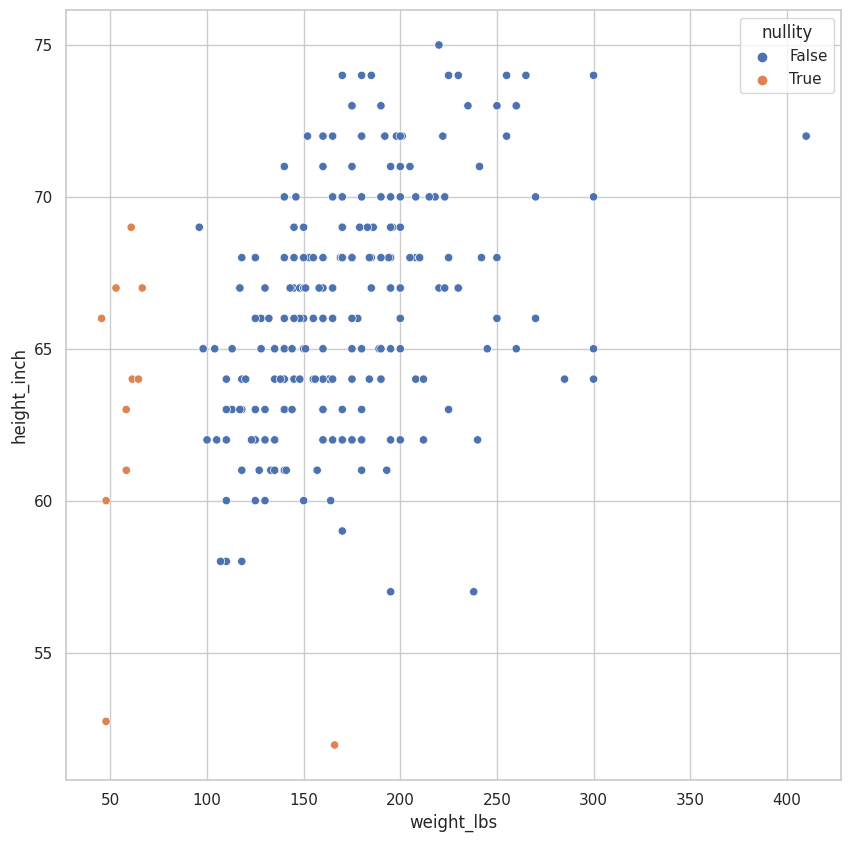
(
riskfactors_df
.select_dtypes(
exclude="category"
)
.pipe(
lambda df: df[df.columns[df.isna().any()]]
)
.missing.bind_shadow_matrix(true_string = True, false_string = False)
.apply(
lambda column: column if "_NA" in column.name else column_fill_with_dummies(column, proportion_below=0.05, jitter = 0.075)
)
.assign(
nullity = lambda df: df.weight_lbs_NA | df.height_inch_NA
)
.pipe(
lambda df: (
sns.scatterplot(
data = df,
x = "weight_lbs",
y = "height_inch",
hue = "nullity"
)
)
)
)
<AxesSubplot:xlabel='weight_lbs', ylabel='height_inch'>
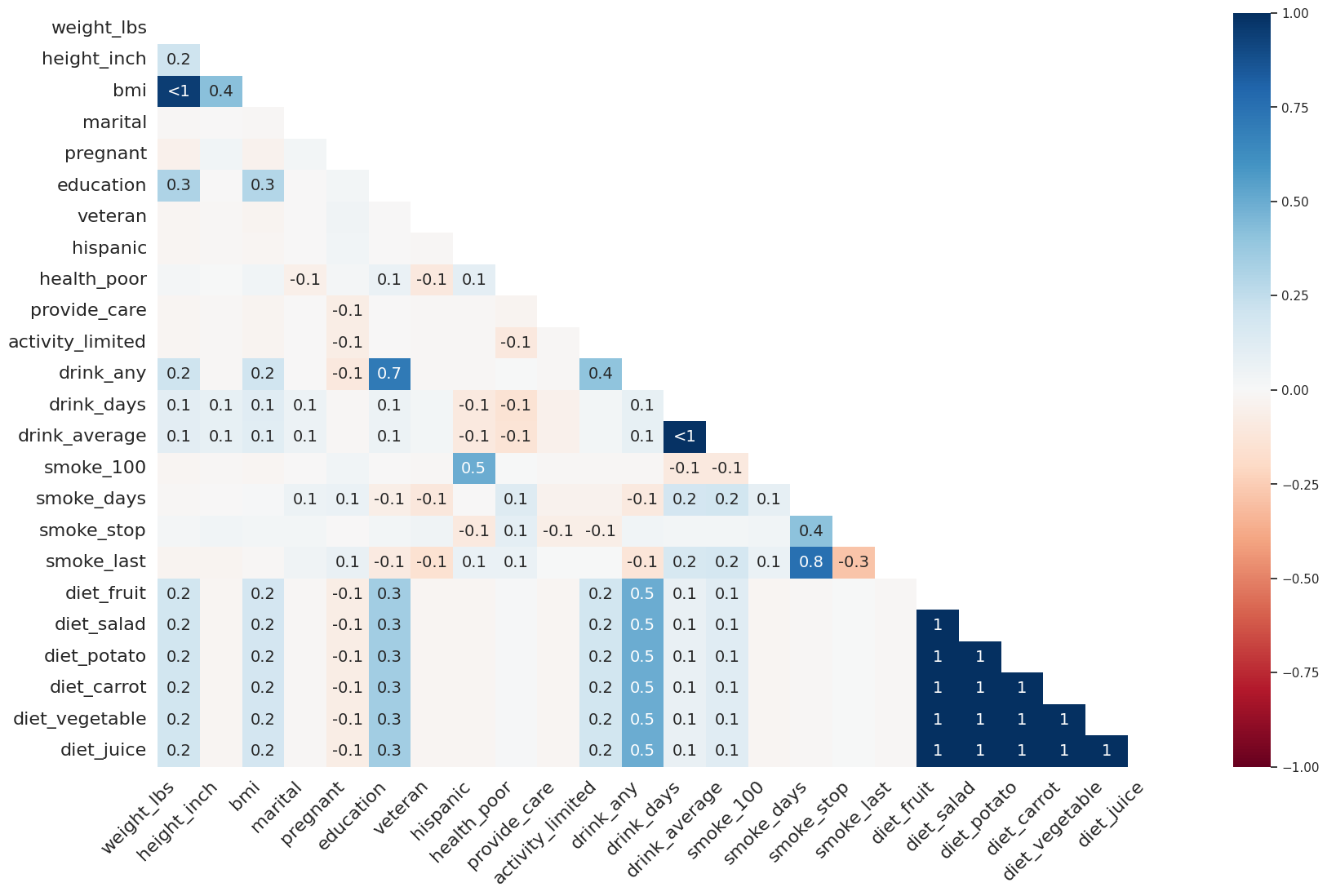
Correlación de nulidad#
missingno.heatmap(
df=riskfactors_df
)
/shared-libs/python3.9/py/lib/python3.9/site-packages/seaborn/matrix.py:306: MatplotlibDeprecationWarning: Auto-removal of grids by pcolor() and pcolormesh() is deprecated since 3.5 and will be removed two minor releases later; please call grid(False) first.
mesh = ax.pcolormesh(self.plot_data, cmap=self.cmap, **kws)
/shared-libs/python3.9/py/lib/python3.9/site-packages/seaborn/matrix.py:316: MatplotlibDeprecationWarning: Auto-removal of grids by pcolor() and pcolormesh() is deprecated since 3.5 and will be removed two minor releases later; please call grid(False) first.
cb = ax.figure.colorbar(mesh, cax, ax, **self.cbar_kws)
<AxesSubplot:>
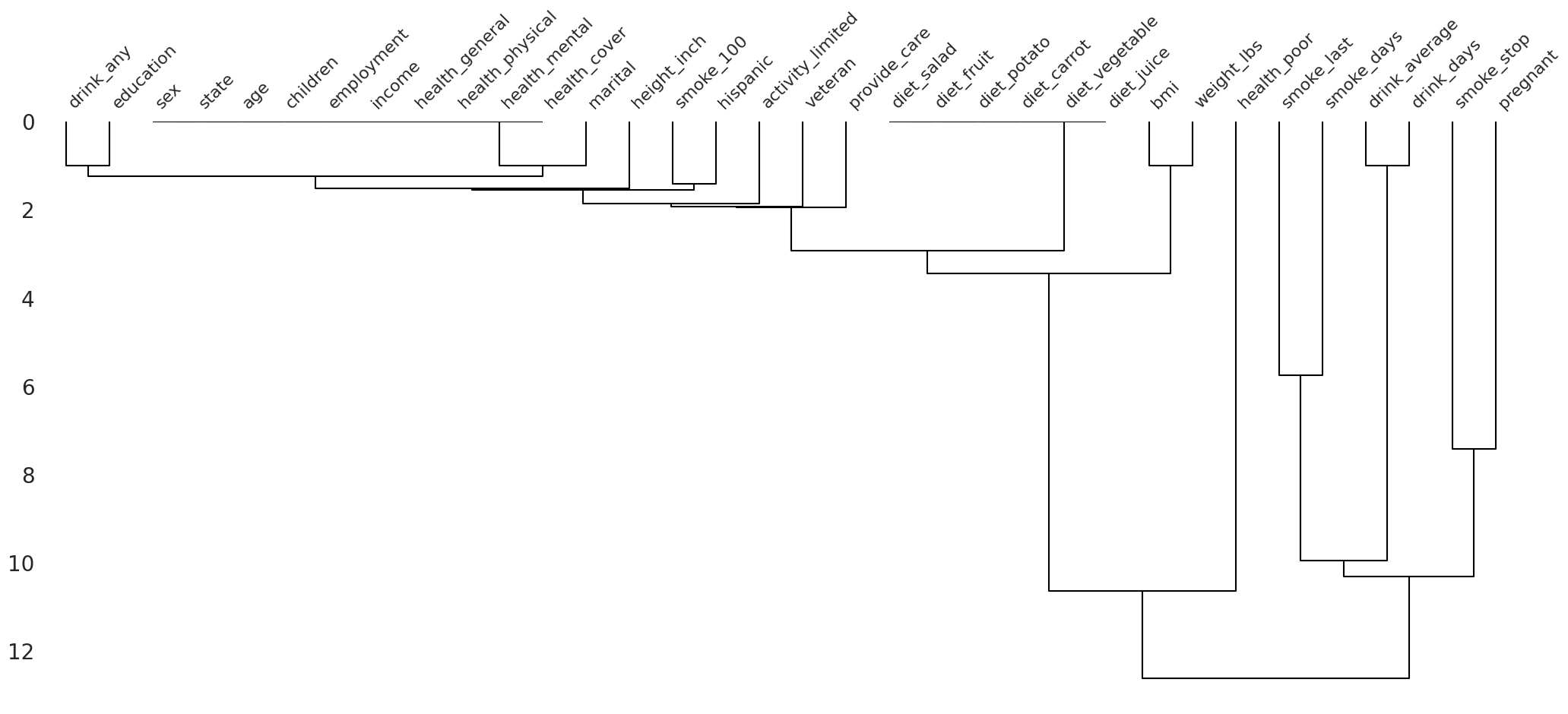
missingno.dendrogram(
df=riskfactors_df
)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:475: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax0.grid(b=False)
<AxesSubplot:>
Eliminación de valores faltantes#
La eliminación de valores faltantes asume que los valores faltantes están perdidos
completamente al azar (MCAR). En cualquier otro caso, realizar una
eliminación de valores faltantes podrá ocasionar sesgos en los
análisis y modelos subsecuentes.
Primero observa el número total de observaciones y variables que tiene tu conjunto de datos.
riskfactors_df.shape
(245, 34)
Pairwise deletion (eliminación por pares)#
Esto consiste en ignorar valores nulos para realziar operaciones como calculos de promedio, conteos, etc
riskfactors_df.weight_lbs.size, riskfactors_df.weight_lbs.count()
(245, 235)
En este caso, en size se cuentan todos los datos. Mientras que en count() solo se cuentan los datos no nulos
Listwise Deletion or Complete Case (Eliminación por lista o caso completo)#
Este procedimiento consiste en borrar todo el registro (o fila) si es que hay un valor faltante
Con base en 1 columna#
(
riskfactors_df
.dropna(
subset=["weight_lbs"],
how="any"
)
)
| state | sex | age | weight_lbs | height_inch | bmi | marital | pregnant | children | education | ... | smoke_100 | smoke_days | smoke_stop | smoke_last | diet_fruit | diet_salad | diet_potato | diet_carrot | diet_vegetable | diet_juice | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 26 | Female | 49 | 190 | 64 | 32.68 | Married | NaN | 0 | 6 | ... | No | NaN | NaN | NaN | 1095 | 261 | 104 | 156 | 521 | 12 |
| 1 | 40 | Female | 48 | 170 | 68 | 25.90 | Divorced | NaN | 0 | 5 | ... | No | NaN | NaN | NaN | 52 | 209 | 52 | 0 | 52 | 0 |
| 2 | 72 | Female | 55 | 163 | 64 | 28.04 | Married | NaN | 0 | 4 | ... | No | NaN | NaN | NaN | 36 | 156 | 52 | 24 | 24 | 24 |
| 3 | 42 | Male | 42 | 230 | 74 | 29.59 | Married | NaN | 1 | 6 | ... | No | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | 32 | Female | 66 | 135 | 62 | 24.74 | Widowed | NaN | 0 | 5 | ... | Yes | Everyday | Yes | NaN | -7 | 261 | 209 | 261 | 365 | 104 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 240 | 10 | Female | 79 | 144 | 63 | 25.56 | Widowed | NaN | 0 | 4 | ... | No | NaN | NaN | NaN | -7 | -7 | -7 | -7 | -7 | -7 |
| 241 | 46 | Male | 45 | 170 | 74 | 21.87 | Divorced | NaN | 2 | 4 | ... | No | NaN | NaN | NaN | 52 | 52 | 52 | 24 | 52 | 24 |
| 242 | 15 | Male | 62 | 175 | 71 | 24.46 | Divorced | NaN | 0 | 6 | ... | Yes | Not@All | NaN | 7 | 365 | 156 | 104 | 52 | 730 | 365 |
| 243 | 34 | Female | 62 | 138 | 64 | 23.74 | Married | NaN | 0 | 4 | ... | Yes | Everyday | No | NaN | 730 | 0 | 24 | 156 | 104 | 0 |
| 244 | 18 | Male | 9 | 200 | 70 | 28.76 | Married | NaN | 0 | 4 | ... | Yes | Not@All | NaN | 7 | 52 | 104 | 52 | 0 | 104 | 104 |
235 rows × 34 columns
Se borraron los registros si weight_lbs esta faltando
Con base en 2 o más columnas#
(
riskfactors_df
.dropna(
subset=["weight_lbs", "height_inch"],
how="any"
)
)
| state | sex | age | weight_lbs | height_inch | bmi | marital | pregnant | children | education | ... | smoke_100 | smoke_days | smoke_stop | smoke_last | diet_fruit | diet_salad | diet_potato | diet_carrot | diet_vegetable | diet_juice | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 26 | Female | 49 | 190 | 64 | 32.68 | Married | NaN | 0 | 6 | ... | No | NaN | NaN | NaN | 1095 | 261 | 104 | 156 | 521 | 12 |
| 1 | 40 | Female | 48 | 170 | 68 | 25.90 | Divorced | NaN | 0 | 5 | ... | No | NaN | NaN | NaN | 52 | 209 | 52 | 0 | 52 | 0 |
| 2 | 72 | Female | 55 | 163 | 64 | 28.04 | Married | NaN | 0 | 4 | ... | No | NaN | NaN | NaN | 36 | 156 | 52 | 24 | 24 | 24 |
| 3 | 42 | Male | 42 | 230 | 74 | 29.59 | Married | NaN | 1 | 6 | ... | No | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | 32 | Female | 66 | 135 | 62 | 24.74 | Widowed | NaN | 0 | 5 | ... | Yes | Everyday | Yes | NaN | -7 | 261 | 209 | 261 | 365 | 104 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 240 | 10 | Female | 79 | 144 | 63 | 25.56 | Widowed | NaN | 0 | 4 | ... | No | NaN | NaN | NaN | -7 | -7 | -7 | -7 | -7 | -7 |
| 241 | 46 | Male | 45 | 170 | 74 | 21.87 | Divorced | NaN | 2 | 4 | ... | No | NaN | NaN | NaN | 52 | 52 | 52 | 24 | 52 | 24 |
| 242 | 15 | Male | 62 | 175 | 71 | 24.46 | Divorced | NaN | 0 | 6 | ... | Yes | Not@All | NaN | 7 | 365 | 156 | 104 | 52 | 730 | 365 |
| 243 | 34 | Female | 62 | 138 | 64 | 23.74 | Married | NaN | 0 | 4 | ... | Yes | Everyday | No | NaN | 730 | 0 | 24 | 156 | 104 | 0 |
| 244 | 18 | Male | 9 | 200 | 70 | 28.76 | Married | NaN | 0 | 4 | ... | Yes | Not@All | NaN | 7 | 52 | 104 | 52 | 0 | 104 | 104 |
234 rows × 34 columns
Se borraron los registros si weight_lbs y height_inch estan faltando
(
riskfactors_df
.dropna(
subset=["weight_lbs", "height_inch"],
how="all"
)
)
| state | sex | age | weight_lbs | height_inch | bmi | marital | pregnant | children | education | ... | smoke_100 | smoke_days | smoke_stop | smoke_last | diet_fruit | diet_salad | diet_potato | diet_carrot | diet_vegetable | diet_juice | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 26 | Female | 49 | 190 | 64 | 32.68 | Married | NaN | 0 | 6 | ... | No | NaN | NaN | NaN | 1095 | 261 | 104 | 156 | 521 | 12 |
| 1 | 40 | Female | 48 | 170 | 68 | 25.90 | Divorced | NaN | 0 | 5 | ... | No | NaN | NaN | NaN | 52 | 209 | 52 | 0 | 52 | 0 |
| 2 | 72 | Female | 55 | 163 | 64 | 28.04 | Married | NaN | 0 | 4 | ... | No | NaN | NaN | NaN | 36 | 156 | 52 | 24 | 24 | 24 |
| 3 | 42 | Male | 42 | 230 | 74 | 29.59 | Married | NaN | 1 | 6 | ... | No | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | 32 | Female | 66 | 135 | 62 | 24.74 | Widowed | NaN | 0 | 5 | ... | Yes | Everyday | Yes | NaN | -7 | 261 | 209 | 261 | 365 | 104 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 240 | 10 | Female | 79 | 144 | 63 | 25.56 | Widowed | NaN | 0 | 4 | ... | No | NaN | NaN | NaN | -7 | -7 | -7 | -7 | -7 | -7 |
| 241 | 46 | Male | 45 | 170 | 74 | 21.87 | Divorced | NaN | 2 | 4 | ... | No | NaN | NaN | NaN | 52 | 52 | 52 | 24 | 52 | 24 |
| 242 | 15 | Male | 62 | 175 | 71 | 24.46 | Divorced | NaN | 0 | 6 | ... | Yes | Not@All | NaN | 7 | 365 | 156 | 104 | 52 | 730 | 365 |
| 243 | 34 | Female | 62 | 138 | 64 | 23.74 | Married | NaN | 0 | 4 | ... | Yes | Everyday | No | NaN | 730 | 0 | 24 | 156 | 104 | 0 |
| 244 | 18 | Male | 9 | 200 | 70 | 28.76 | Married | NaN | 0 | 4 | ... | Yes | Not@All | NaN | 7 | 52 | 104 | 52 | 0 | 104 | 104 |
244 rows × 34 columns
Se borraron los registros si weight_lbs o height_inch estan faltando
Representación gráfica tras la eliminación de los valores faltantes#
(
riskfactors_df
.dropna(
subset=["weight_lbs", "height_inch"],
how="any"
)
.select_columns(["weight_lbs", "height_inch"])
.pipe(missingno.matrix)
)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:73: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax0.grid(b=False)
/root/venv/lib/python3.9/site-packages/missingno/missingno.py:142: MatplotlibDeprecationWarning: The 'b' parameter of grid() has been renamed 'visible' since Matplotlib 3.5; support for the old name will be dropped two minor releases later.
ax1.grid(b=False)
<AxesSubplot:>
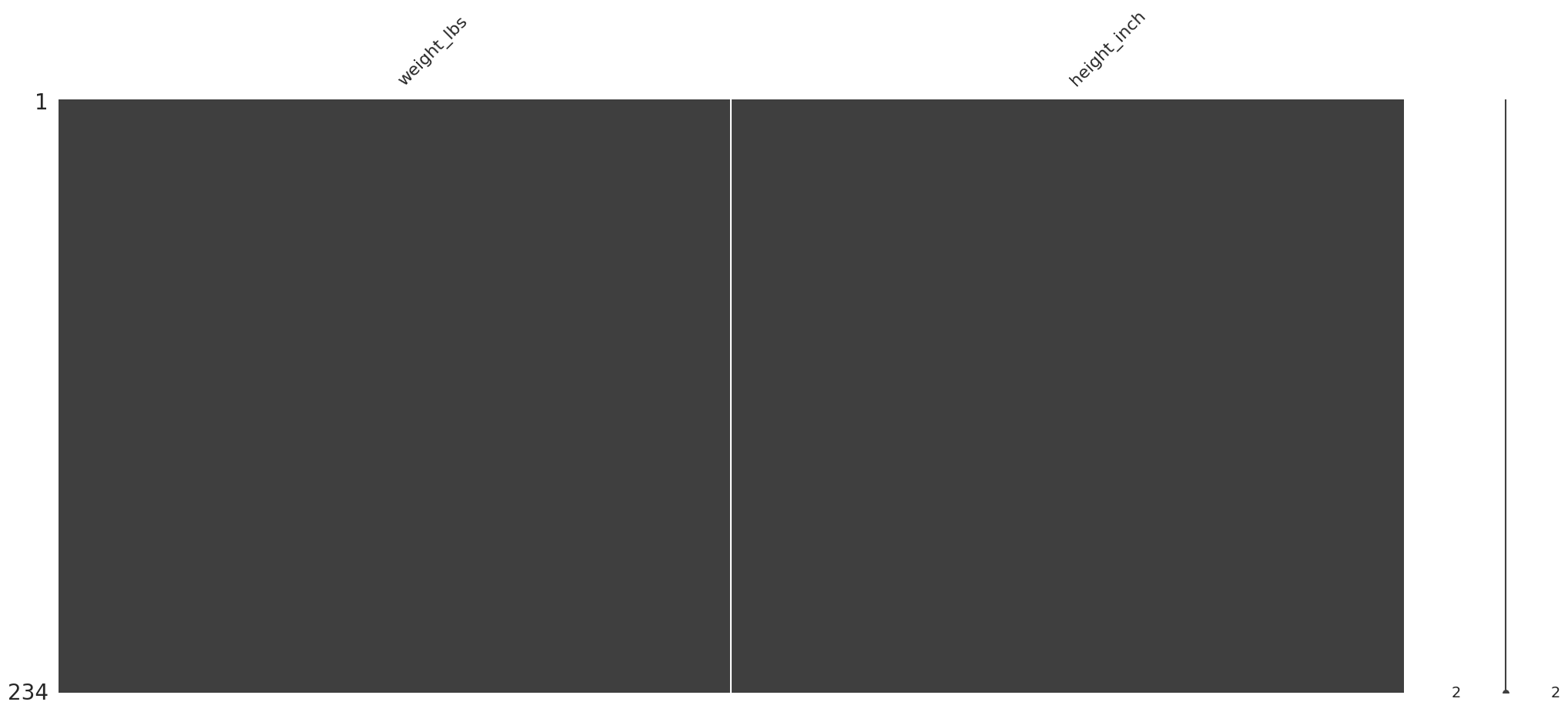
Se confirma en el grafico que no faltan datos
Imputación básica de valores faltantes#
Imputacion es reemplazar los datos faltantes por datos con sentido, puedes basarte en:
algun estadistico que venga de los datos
un valor por contexto
un valor extraido de un modelo de machine learning
Imputación con base en el contexto#
implicit_to_explicit_df = pd.DataFrame(
data={
"name": ["lynn", np.nan, "zelda", np.nan, "shadowsong", np.nan],
"time": ["morning", "afternoon", "morning", "afternoon", "morning", "afternoon",],
"value": [350, 310, 320, 350, 310, 320]
}
)
implicit_to_explicit_df
| name | time | value | |
|---|---|---|---|
| 0 | lynn | morning | 350 |
| 1 | NaN | afternoon | 310 |
| 2 | zelda | morning | 320 |
| 3 | NaN | afternoon | 350 |
| 4 | shadowsong | morning | 310 |
| 5 | NaN | afternoon | 320 |
En este caso, es obvio que hay que poner el mismo nombre de la fila previa en el valor faltante, eso es contexto.
Como se rellena de manera rapida?
implicit_to_explicit_df.ffill()
| name | time | value | |
|---|---|---|---|
| 0 | lynn | morning | 350 |
| 1 | lynn | afternoon | 310 |
| 2 | zelda | morning | 320 |
| 3 | zelda | afternoon | 350 |
| 4 | shadowsong | morning | 310 |
| 5 | shadowsong | afternoon | 320 |
Imputación de un único valor#
(
riskfactors_df
.select_columns("weight_lbs", "height_inch", "bmi")
.missing.bind_shadow_matrix(true_string = True, false_string = False)
.apply(
axis = "rows",
func = lambda column : column.fillna(column.mean()) if "_NA" not in column.name else column
# Basicamente se acaba de llenar el valor faltante con el promedio de su respectiva columna
)
)
| weight_lbs | height_inch | bmi | weight_lbs_NA | height_inch_NA | bmi_NA | |
|---|---|---|---|---|---|---|
| 0 | 190.0 | 64.0 | 32.68 | False | False | False |
| 1 | 170.0 | 68.0 | 25.90 | False | False | False |
| 2 | 163.0 | 64.0 | 28.04 | False | False | False |
| 3 | 230.0 | 74.0 | 29.59 | False | False | False |
| 4 | 135.0 | 62.0 | 24.74 | False | False | False |
| ... | ... | ... | ... | ... | ... | ... |
| 240 | 144.0 | 63.0 | 25.56 | False | False | False |
| 241 | 170.0 | 74.0 | 21.87 | False | False | False |
| 242 | 175.0 | 71.0 | 24.46 | False | False | False |
| 243 | 138.0 | 64.0 | 23.74 | False | False | False |
| 244 | 200.0 | 70.0 | 28.76 | False | False | False |
245 rows × 6 columns
Basicamente se acaba de llenar el valor faltante con el promedio de su respectiva columna
visualizando ese procedimiento:
(
riskfactors_df
.select_columns("weight_lbs", "height_inch", "bmi")
.missing.bind_shadow_matrix(true_string = True, false_string = False)
.apply(
axis = "rows",
func = lambda column : column.fillna(column.mean()) if "_NA" not in column.name else column
# Basicamente se acaba de llenar el valor faltante con el promedio de su respectiva columna
)
.pipe(
lambda df: (
sns.displot(
data=df,
x="weight_lbs",
hue="weight_lbs_NA"
)
)
)
)
<seaborn.axisgrid.FacetGrid at 0x7f3faf6c0ca0>
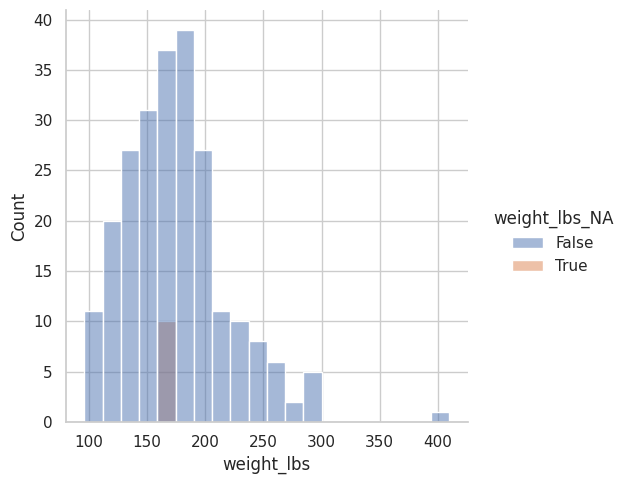
Se confirma que los valores faltantes ahora corresponden al promedio.
Scatterplot con valores imputados#
(
riskfactors_df
.select_columns("weight_lbs", "height_inch", "bmi")
.missing.bind_shadow_matrix(true_string = True, false_string = False)
.apply(
axis = "rows",
func = lambda column : column.fillna(column.mean()) if "_NA" not in column.name else column
)
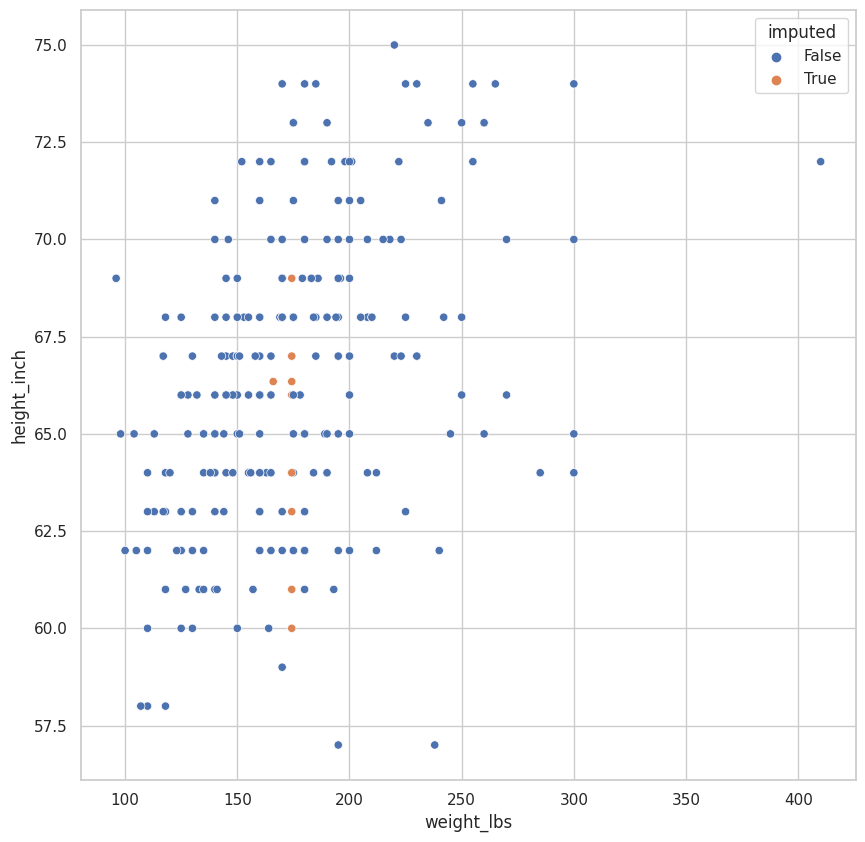
.assign(
imputed=lambda df: df.weight_lbs_NA | df.height_inch_NA
)
.pipe(
lambda df: (
sns.scatterplot(
data=df,
x="weight_lbs",
y="height_inch",
hue="imputed"
)
)
)
)
<AxesSubplot:xlabel='weight_lbs', ylabel='height_inch'>
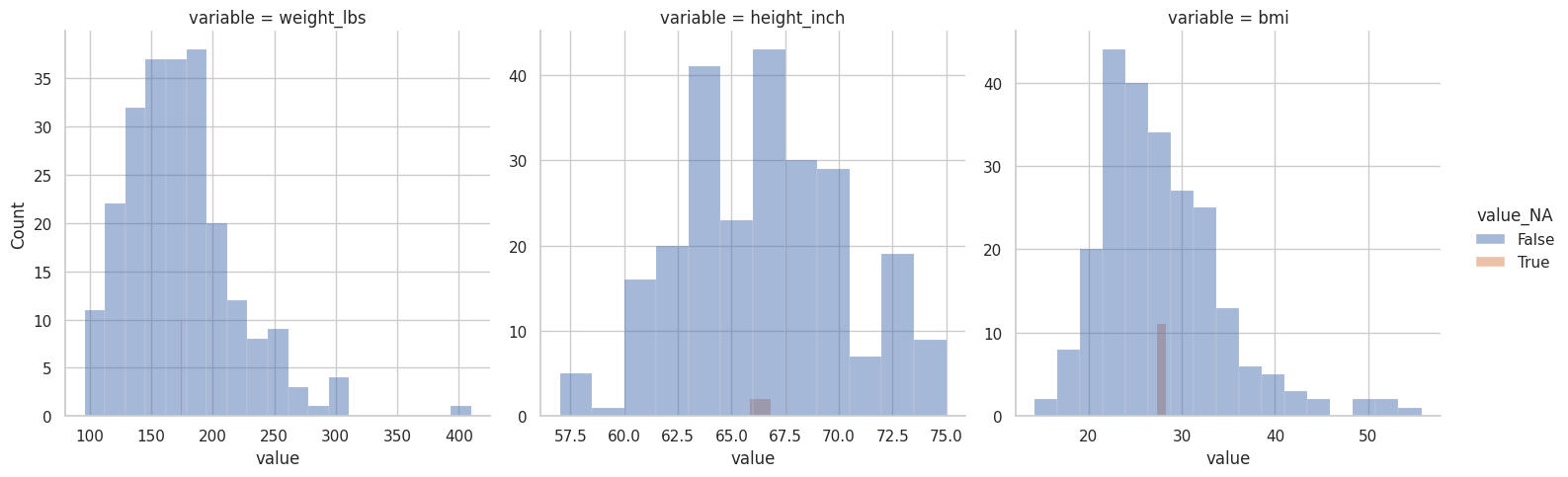
(
riskfactors_df
.select_columns("weight_lbs", "height_inch", "bmi")
.missing.bind_shadow_matrix(true_string = True, false_string = False)
.apply(
axis = "rows",
func = lambda column : column.fillna(column.mean()) if "_NA" not in column.name else column
)
.pivot_longer(
index="*_NA"
)
.pivot_longer(
index=["variable", "value"],
names_to = "variable_NA",
values_to = "value_NA"
)
.assign(
valid=lambda df:df.apply(axis="columns", func = lambda column: column.variable in column.variable_NA)
)
.query("valid")
.pipe(
lambda df: (
sns.displot(
data=df,
x="value",
hue="value_NA",
col="variable",
common_bins=False,
facet_kws={
"sharex": False,
"sharey": False
}
)
)
)
)
<seaborn.axisgrid.FacetGrid at 0x7f3fbf26bd60>